Method for detecting deletion and/or duplication of exons in DMD gene
An exon deletion and exon technology, which is applied in biochemical equipment and methods, and the determination/inspection of microorganisms, can solve the problems of low throughput and poor efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0071] In this example, the sequences and names of several important primers are shown in Table 1.
[0072] Table 1
[0073]
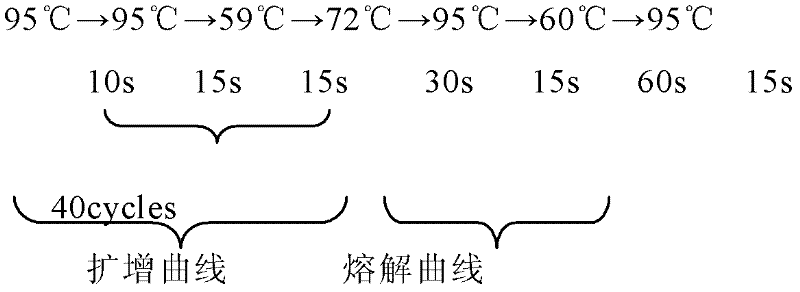
[0074]The first primer (SEQ ID NO.1) and the second primer (SEQ ID NO.2) amplify the DNA double-stranded nucleic acid fragment with the adapter to obtain the first PCR amplification product, the first primer and the second primer There is an adapter binding region corresponding to the primer binding region of the adapter, and a sequencing probe binding region located outside the adapter binding region. The function of blocking molecule 1 (SEQ ID NO.3) and blocking molecule 2 (SEQ ID NO.4) is to complement the linker during sequence capture and avoid capturing non-specific sequences. The function of the third primer (SEQ ID NO.5) and the fourth primer (SEQ ID NO.6) is to amplify the captured specific DNA fragments in large quantities for the next step of sequencing.
[0075] This study recruited 1 DMD female carrier and 4 normal females, who signe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com