Probes for detection of polymorphisms of c-kit gene, and use thereof
A technology of polymorphism detection and polymorphism, which is applied in the determination/inspection of microorganisms, material analysis through observation of the influence of chemical indicators, organic chemistry, etc., can solve problems such as costing a lot of labor and being impractical
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
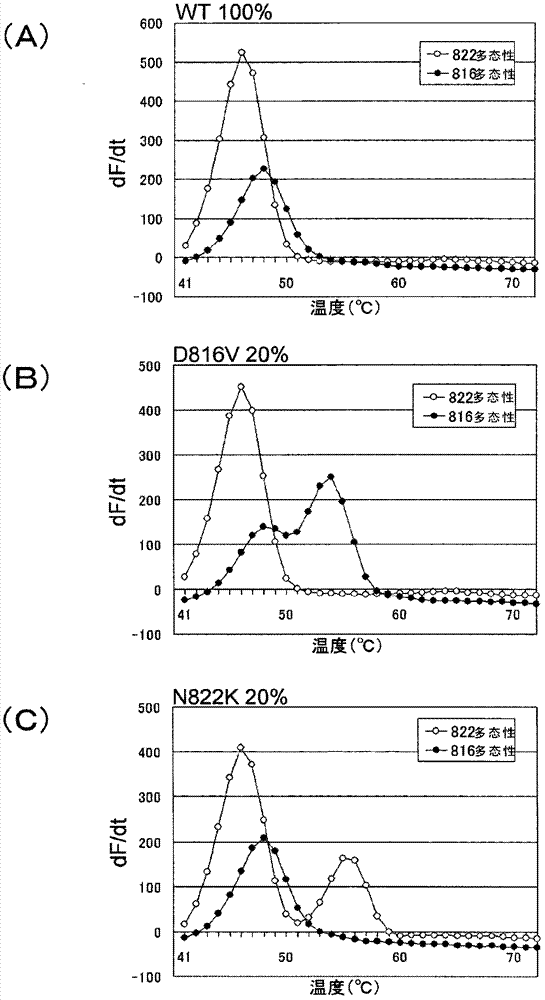
[0221] In this example, Tm analysis was performed to detect the polymorphism of the c-kit gene under the condition that the wild type plasmid and the mutant plasmid coexisted.
[0222] As a partial sequence of the c-kit gene, a wild-type plasmid (WT) inserted with an oligonucleotide (SEQ ID NO: 23) having the D816 wild type and N822 wild type, and an oligonucleotide inserted with the D816V mutant type and the N822 wild type were prepared. D816V mutant plasmid (D816V) of oligonucleotide (SEQ ID NO: 24), N822K mutant plasmid (N822K) inserted with oligonucleotide (SEQ ID NO: 25) having D816 wild type and N822K mutant type. In the wild-type plasmid (WT), the base (w) of base number 108 in the base sequence shown in SEQ ID NO: 1 is adenine (a), and the base (d) of base number 127 is Thymine (t). In the D816V mutant plasmid (D816V), the base number 108 (w) in the base sequence shown in sequence number 1 is thymine (t), and the base number 127 (d) is thymine (t). In the N822K muta...
Embodiment 2
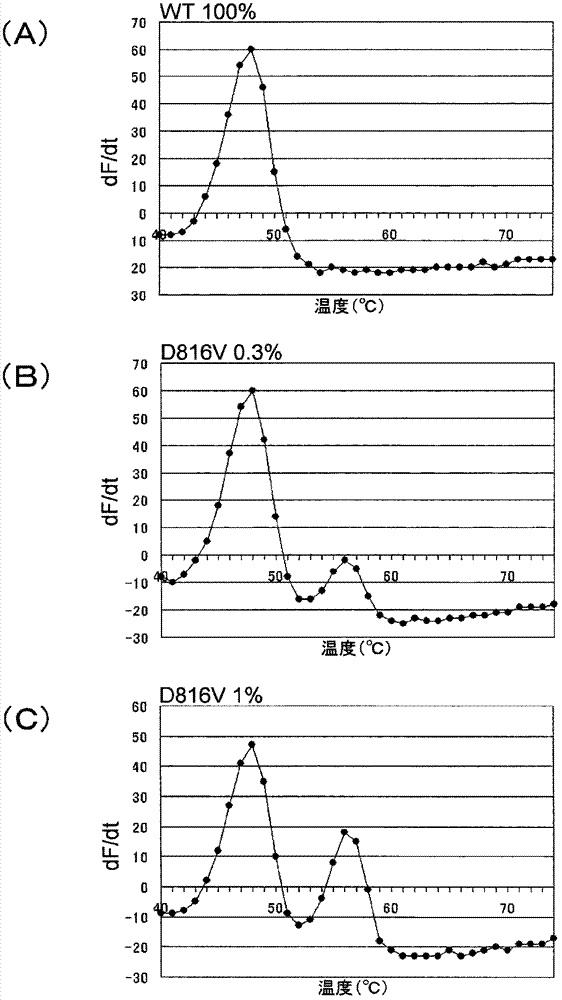
[0252] In this example, Tm analysis was performed in the coexistence of wild-type and mutant plasmids to detect the 816 polymorphism of the c-kit gene.
[0253] The D816V mutant plasmid (D816V) inserted with the oligonucleotides having the wild-type plasmid (WT) and the D816V mutant type described in Example 1 was mixed at a predetermined ratio as shown below to prepare three kinds of plasmid samples. For each 1 μL of plasmid sample, 2×10 4 copies / μL.
[0254] [Table 4]
[0255]
[0256] PCR and Tm analysis were performed in the same manner as in Example 1 except that 50 μL of the PCR reaction solution in Table 4 below and the following PCR and Tm analysis conditions were used. The PCR and Tm analysis conditions are as follows: 95°C for 1 second and 61°C for 15 seconds as a cycle, repeated 50 cycles after 95°C for 60 seconds, and 95°C for 1 second and 40°C for 60 seconds , and then set the rate of temperature rise to 1° C. / 3 seconds, heating from 40° C. to 75° C., and a ...
Embodiment 3
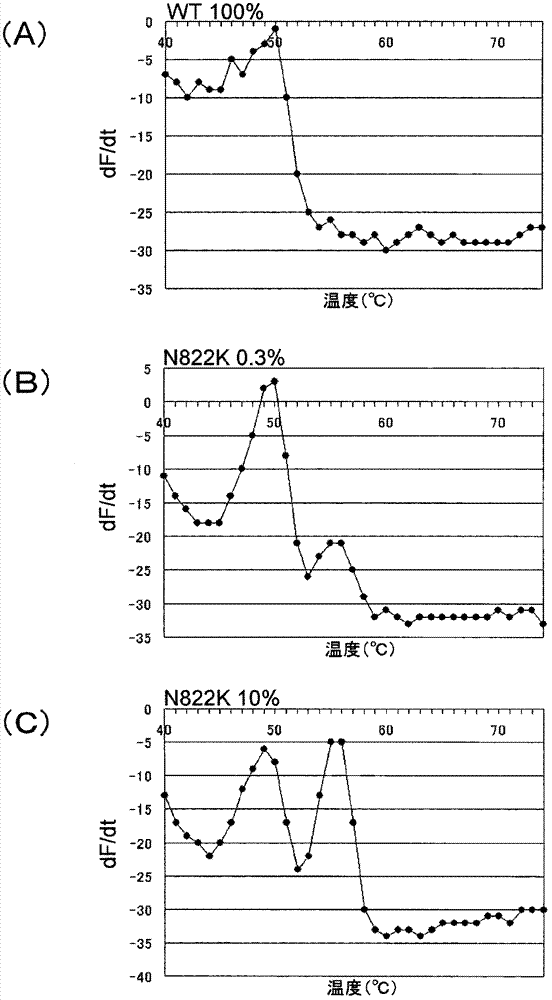
[0274] In this example, Tm analysis was performed to detect the 822 polymorphism of the c-kit gene under the condition that the wild-type plasmid and the mutant plasmid co-existed.
[0275] The N822K mutant plasmid (N822K) inserted with the oligonucleotides having the wild-type plasmid (WT) and the N822K mutant type described in Example 1 was mixed at a predetermined ratio shown below to prepare three kinds of plasmid samples. 1 × 10 per 1 μL of each plasmid sample 4 copies / μL.
[0276] [Table 6]
[0277]
[0278]
[0279] PCR and Tm analysis were performed in the same manner as in Example 2, except that the above-mentioned plasmid samples, the primers and probes shown below, and the following PCR conditions were used. The PCR conditions are 95°C for 60 seconds, 95°C for 1 second and 63°C for 15 seconds as a cycle, repeating 50 cycles, then 95°C for 1 second, and 40°C for 1 second. 60 seconds.
[0280] The sequences of the wild-type F primer and the mutant-type F pri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com