Liver cancer diagnostic marker composed of blood plasma microRNA (micro ribonucleic acid) and new method for diagnosing liver cancer
A diagnostic marker, hsa-mir-21 technology, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of limited number of microRNA, lack of validation, small sample size, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0165] Example 1: Collection and preparation of plasma samples:
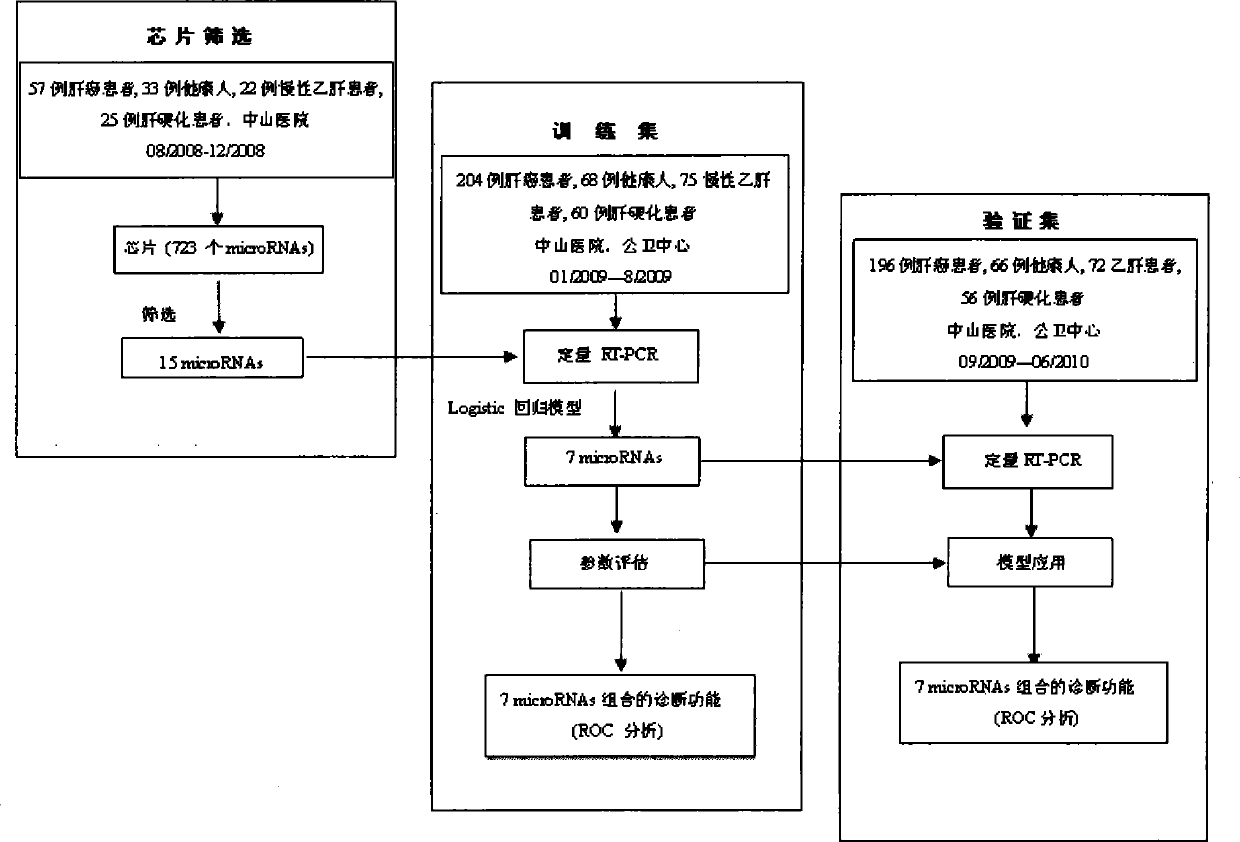
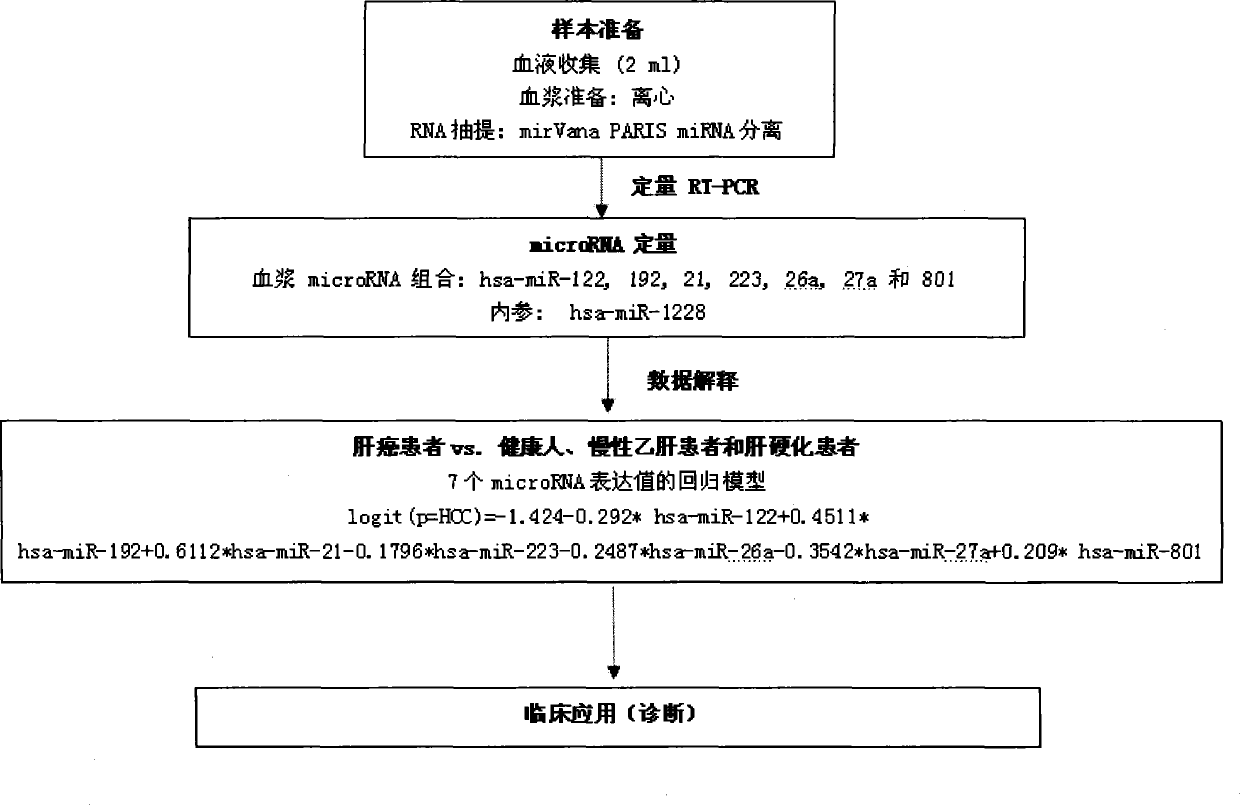
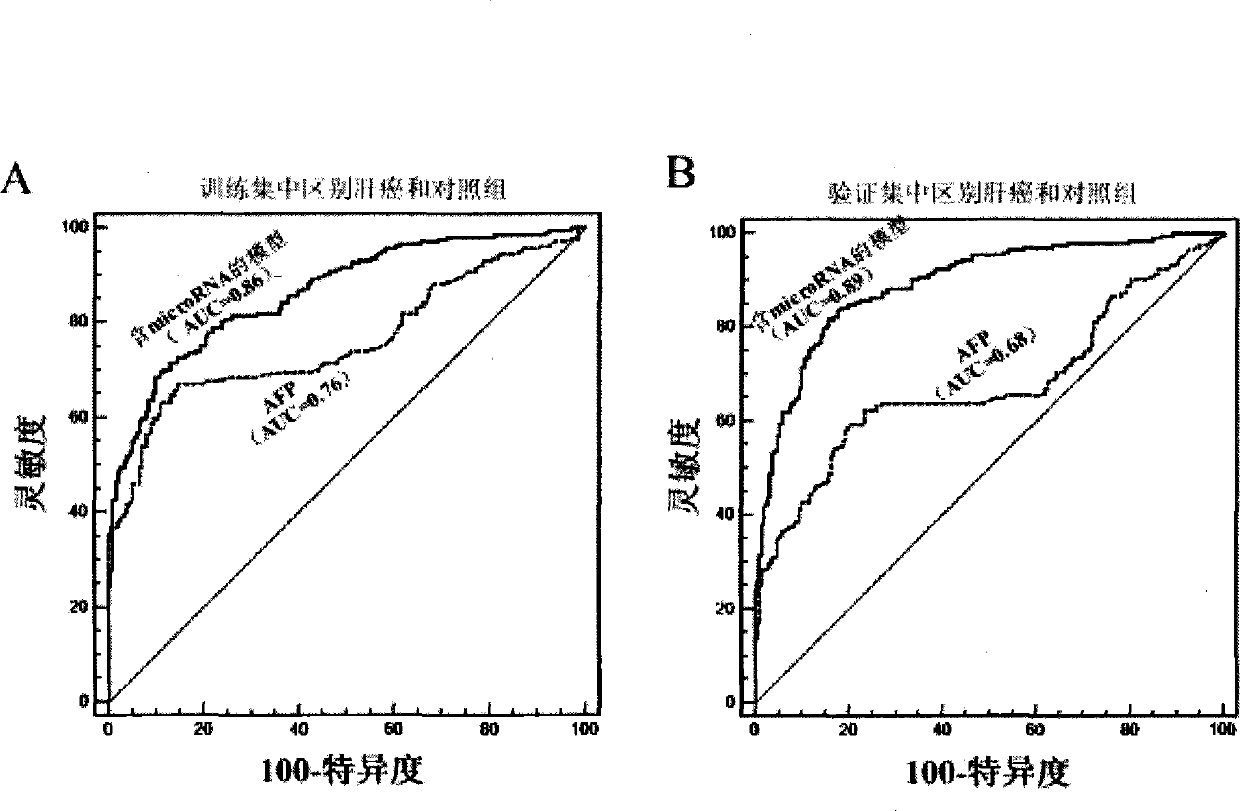
[0166] The experiments in this invention have been approved by the local ethics committee and informed consent was obtained from all patients. The experimental design of the screening stage, training stage and verification stage of microRNA biomarkers in the present invention is as follows: figure 1 shown. The main method steps for identifying plasma samples from HCC patients with the recommended plasma microRNA panels are as follows: figure 2 shown.
[0167] Between August 2008 and June 2010, 934 met the eligibility criteria ( figure 2 ) blood samples were collected in advance from Shanghai Zhongshan Hospital and Public Health Center. These samples were obtained from 167 healthy donors (healthy group), 169 patients with chronic hepatitis B (CHB group), 141 patients with cirrhosis after HBV infection (cirrhosis group), and 457 HCC patients associated with HBV infection (HCC group) . These samples were di...
Embodiment 2
[0182] Embodiment 2: microRNA microarray analysis:
[0183] Qualitative analysis of microRNA in specific plasma samples was performed using the Agilent microRNA microarray platform (Agilent microRNA microarray platform, Agilent Technologies, Santa Clara, CA, USA). The microarray contains probes for 723 human microRNAs from Sanger database v.10.1. Total RNA (100 ng) from any one of the 137 plasma samples was spiked into single-color CY3 for labeling. Scan the gene chip with XDR scanner (PMT100, PMT5), and perform labeling and hybridization according to the protocol of Agilent microRNA microarray system. Microarray image information was converted into spot intensity values by Feature Extraction Software Rev. 9.5.3 (Agilent Technologies, Santa Clara, CA). The signal after background removal was normalized with the stable internal reference hsa-miR-1228. Then, a base-2 log transformation is performed. An unreliable sample with an inter-slice variation coefficient (CV) of mor...
Embodiment 3
[0186] Example 3: Microarray data of 137 samples
[0187] The expressions of the 15 candidate microRNAs used for further verification in the microarray analysis are shown in Table 5, and the microRNAs meeting the selection criteria are indicated in bold.
[0188] table 5
[0189] Expression of 15 candidate microRNAs in microarray
[0190]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com