Building method of two-level fitting quantitative structure-activity relationship (QSAR) model for forecasting compound activity
A construction method and a technology for fitting models, which are applied in the field of biomedical information, can solve problems such as model performance degradation, modeling failure, and non-convergence, and achieve the effects of fewer independent variables, preventing modeling failure, and easy interpretation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
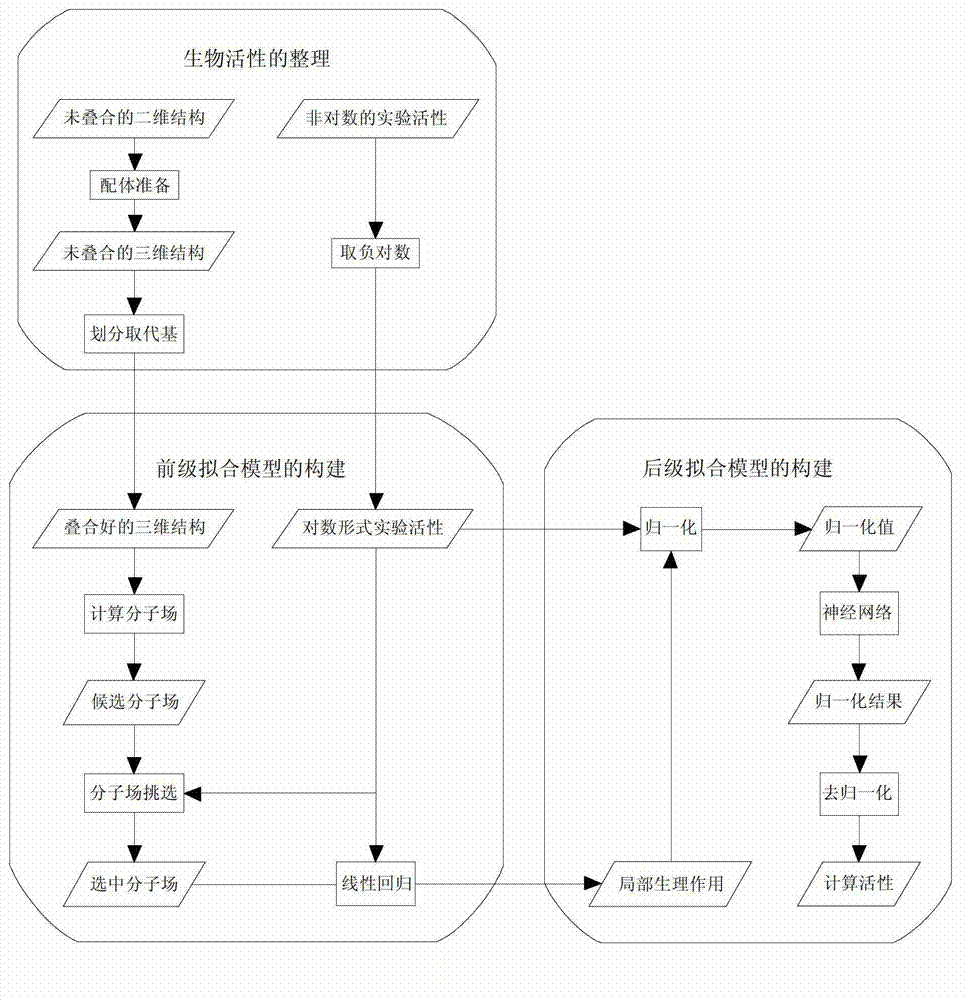
[0039] Such as figure 1 As shown, the linear regression-neural network of the present embodiment fits the QSAR model in two stages before and after, and its construction steps are as follows:
[0040] 1) Finishing of biological activity
[0041] In order to ensure the statistical effect, 35 pyrazole compounds with p38 kinase inhibitory rate were taken as the training set S 1 , convert its inhibition rate α into logarithmic form: Y 1 =LgBio=-lg(α -1 -1). Y 1 =LgBio is the dependent variable used in subsequent modeling. Sybyl analysis software was used to test the two-dimensional structure of the compound, and the three-dimensional structure of the compound that passed the test was generated.
[0042] 2) Construction of pre-fitting model
[0043] The training set compound S 1 Import the molecular table S1.tbl of Sybyl software, in the Topomer CoMFA module, for the training set S 1 The substituents of the compounds are divided. On the one hand, the division of substituent...
Embodiment 2
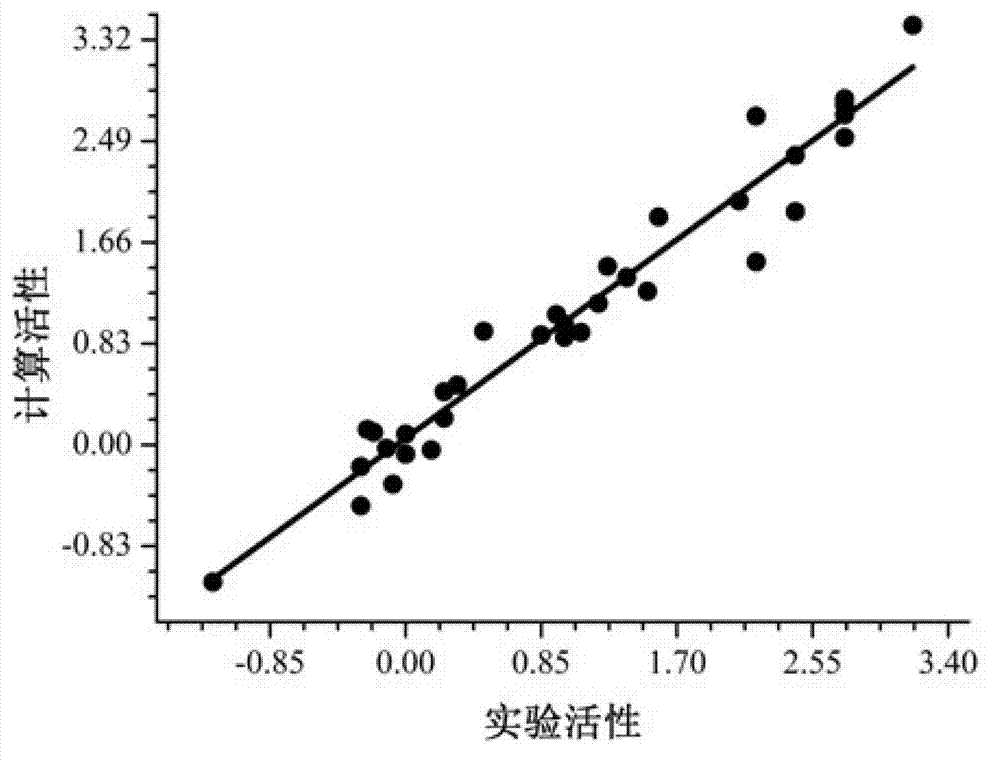
[0047] This embodiment is to measure the goodness of fit, and compare the M 1 -M 2 Two-level model with M 1 The goodness of fit of the single-level model, the specific steps are as follows:
[0048] 1) Variable naming
[0049] will model M 1 For the training set S 1 The calculated activity of the compound is named Y 2 .
[0050] will model M 2 For the training set S 1 The calculated activity of the compound is named Y 3 .
[0051] 2) Export spreadsheet file
[0052] The Sybyl molecule forms S 1 The two columns LgBio and Pre_LgBio in .tbl are exported as S 1 _M 1 .csv file, then converted to S 1 _M 1 .xls file. The above LgBio is Y 1 , Pre_LgBio is Y 2 .
[0053] Using the same method, export M2 from the SPSS Clementine software to the training set compound S 1 The computational activity of , saved as S 1 _M 2 .xls file; where, S 1 _M 2 The .xls file contains the variable Y 1 and Y 3 .
[0054] 3) Calculate the square of the correlation coefficient an...
Embodiment 3
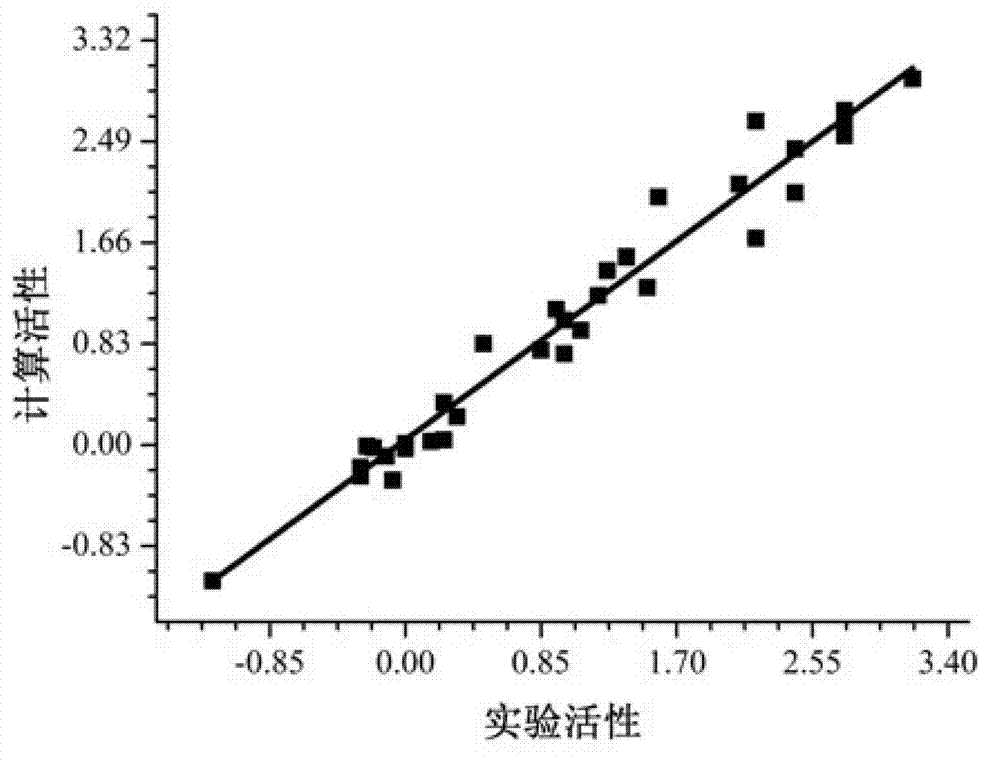
[0059] This embodiment is to measure the predictive performance, compare the above-mentioned embodiment 1 built M 1 -M 2 Two-level model with M 1 The prediction performance of the single-level model, the specific steps are as follows:
[0060] 1) Collation of p38 kinase inhibitory activity
[0061] Take 35 non-training sets S 1 Pyrazole compounds of elements form a test set S 2 , whose p38 kinase inhibitory activity is denoted as Y 4 . The test set S 2 The 35 pyrazole compounds are made into Sybyl molecular form S 2 .tbl, Y 4 Specified as dependent variable (indicated as LgBio in the S2.tbl molecule sheet).
[0062] 2) Determination of the predictive performance of the single-level model M1
[0063] In the TopomerCoMFA module of Sybyl software, the predicted molecular form S 2 .p38 kinase inhibitory activity of tbl, the result is marked as Y 5 (at S 2 Indicated as Pre_LgBio in the .tbl molecule form). During the prediction process, the local physiological effects...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com