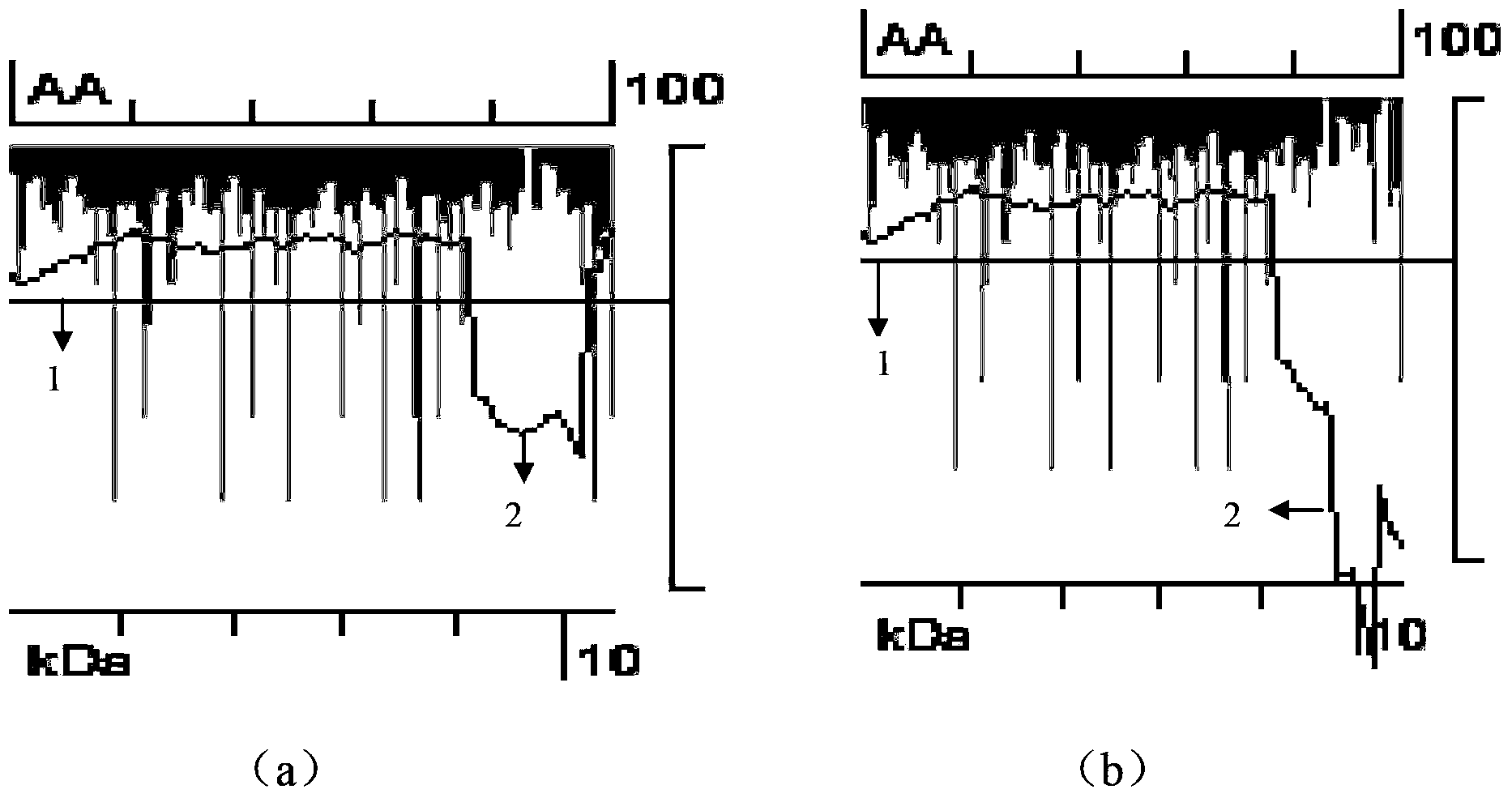
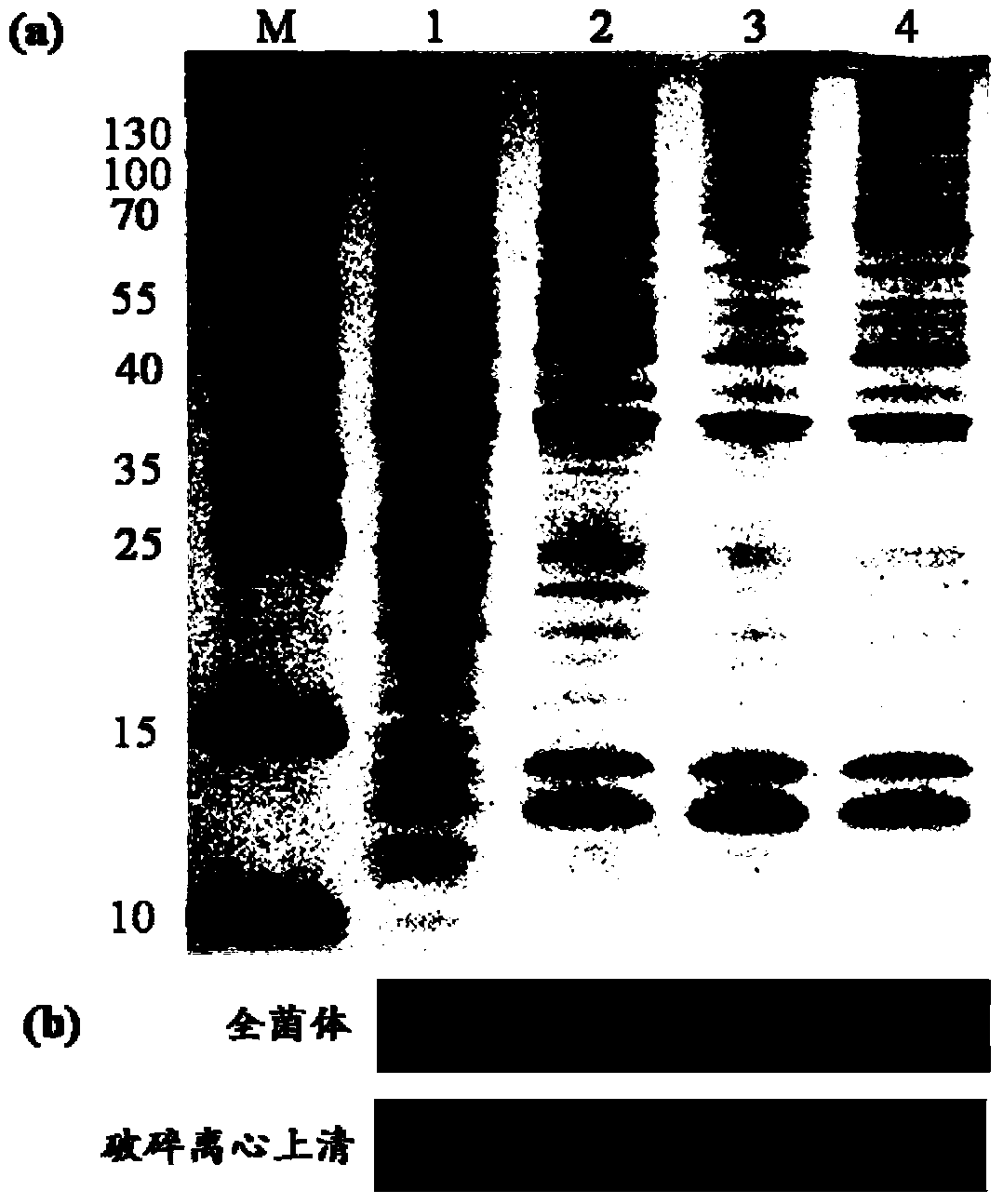
Method for improving protein soluble expression through rational translation pause sequence redesigning
A technology for translational pause sequences and proteins, which is applied in the field of improving protein soluble expression through rational redesign of translational pause sequences, and can solve the problems of protein misfolding and aggregation, slowness, and inability to obtain soluble proteins.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Example 1 CVN codon optimization and expression vector construction
[0071] Using the translation pause theory, optimize the nucleotide sequence encoding CVN protein, rationally design the translation pause site, and carry out synonymous mutations on the nucleotide sequence of CVN by mutation PCR technology.
[0072] The coding nucleotide sequence of CVN protein is as follows:
[0073] CTTGGTAAATTCTCCCAGACCTGCTACAACTCCGCTATCCAGGGTTCTGTTCTGACCTCTACCTGCGAACGTACCAACGGTGGTTACAACACCTCCTCTATCGACCTGAACTCCGTTATCGAAAACGTTGACGGTTCTCTGAAATGGCAGCCGTCTAACTTCATCGAAACCTGCCGTAACACCCAGCTGGCTGGTTCCTCTGAACTGGCTGCTGAATGCAAAACCCGTGCTCAGCAGTTCGTTTCTACCAAAATCAACCTGGACGACCACATCGCTAACATAGACGGTACACTAAAATACGAA。
[0074] (1) Design the nucleotide sequence encoding the CVN mutant by translation pause theory:
[0075] 1) According to the protein structure, determine where the translation pause site should exist
[0076] ① For multi-domain proteins, the translation pause site is designed within: A...
Embodiment 2
[0112] (1) Preparation of expression strains:
[0113] ① Preparation of Escherichia coli BL21 (DE3) competent cells: For the preparation process, refer to the third edition of "Molecular Cloning Experiment Guide"; [US] J. Sambrook, translated by Huang Peitang.
[0114] ②Transform the expression vectors pET-28b-CVN, pET-28b-CVN-M1 and pET-28b-CVN-M2 into Escherichia coli BL21 (DE3) competent cells: see the third part of "Molecular Cloning Experiment Guide" for details on the transformation process. Edition; [US] J. Sambrook, translated by Huang Peitang.
[0115] (2) Induced expression and solubility analysis of CVN and its mutants
[0116] ① Inoculate the expression strains CVN-BL21, CVN-M1-BL21 and CVN-M2-BL21 obtained in step (1) into 20 mL of LB medium containing 50 μg / mL kanamycin content, culture at 37 °C and 180 rpm, When OD 600 When = 0.8, add IPTG, the final concentration is 1mM, after induction expression at 37℃ for 4h, measure the OD 600 , to ensure the volume of ...
Embodiment 3
[0123] Study on Anti-Influenza Virus A / HK / 8 / 68(H3N2) Activity of CVN
[0124] Dog kidney epithelial cells MDCK cells (ATCC, USA) were digested with 0.25% trypsin solution by weight and volume, and then mixed with 2.5×10 5 Add the cells / well into a 96-well cell culture plate (MEM medium, containing 10% calf serum by volume), discard the growth medium after the cells grow into a monolayer, and use the maintenance medium (MEM medium, without calf serum ) respectively prepare the drug CVN protein (preserved in the laboratory and can be prepared according to the steps disclosed in the publication number CN101638435 and titled "A Mutant of Cyanobacterial Virus Protein N, Its Modified Derivatives and Its Application") and Example 2. CVN mutant protein was diluted into 6 serial concentrations (100, 50, 25, 12.5, 6.25, 3.125 μM), and 3 replicate wells were set up for each dilution, and a normal cell control group was set at the same time, positive drug (ribavirin) Control group, 37℃, ...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap