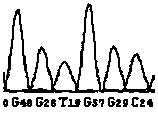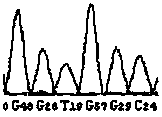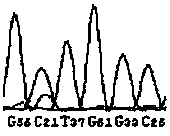Preparation method for nucleic acid mass spectrum for detecting mutation of colorectal cancer K-ras gene and product for detecting mutation of colorectal cancer K-ras gene
A gene and nucleic acid technology, applied in the field of detecting codons 12 and 13 of a K-ras gene and its related products, can solve the problems of high cost, long time, cumbersome steps, etc., achieve high accuracy, high sensitivity, reduce The effect of the cost of consumables
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] Embodiment 1, primer design.
[0082] For the K-ras gene DNA fragment, design corresponding specific PCR primer core sequences (SEQ ID No: 1 and SEQ ID No: 2), the pair of primers can amplify the K-ras gene coding sequence, including 12 and 13 codons 83bp within.
[0083] SEQ ID No: 1: 5'-AAGGCCTGCTGAAAATGACTG-3'
[0084] SEQ ID No: 2: 5'-TAGCTGTATCGTCAAGGCACTCT-3'
[0085] Their amplified sequence SEQ ID NO:3 (83bp) is as follows (the underlined region is codons 12 and 13, and only the wild type is listed):
[0086] SEQ ID No: 3: AAGGCCTGCTGAAAATGACTGAATATAAACTTGTGGTAGTTGGAGCT G GTGGC GTAGGCAAGAGTGCCTTGACGATACAGCTA
[0087] In order to allow reverse transcriptase to recognize PCR products and prevent PCR primers from entering the detection window of the mass spectrometer and interfering with the detection effect, it is also necessary to add specific sequences (lowercase letters) to the 5' ends of SEQ ID No: 1 and SEQ ID No: 2. Therefore, the actually synthesized...
Embodiment 2
[0093] Embodiment 2, sample preparation.
[0094] Through molecular cloning techniques such as plasmid construction and site-directed mutagenesis, a total of 8 plasmids containing K-ras gene wild type and 7 kinds of mutations were constructed, respectively samples WT (wild type), 12A (12 codons GGT>GCT) , 12C (12 codon GGT>TGT), 12D (12 codon GGT>GAT), 12R (12 codon GGT>CGT), 12S (12 codon GGT>AGT), 12V (12 codon GGT>GTT) , 13D (13 codon GGC>GAC). This operation adopts the basic techniques commonly used in molecular biology and will not be repeated here.
[0095] Clinical colorectal cancer patient 1, when the tumor tissue was surgically removed, a fresh tissue block (soybean size) was taken, and the patient's tumor was extracted with a blood / cell / tissue genome extraction kit (Tiangen Biochemical Technology (Beijing) Co., Ltd., DP304) Genomic DNA, designated as sample 1, was reserved at -20°C.
[0096] For patient 2 after colorectal cancer surgery, venous blood was collect...
Embodiment 3
[0099] Example 3. Difference analysis and verification of nuclease digestion fingerprints of wild-type and mutant nuclease-cut fingerprints of codons 12 and 13 of K-ras gene.
[0100] By theoretically analyzing the sequences of the wild type and the mutant type, the wild type and the mutant type will produce different nuclease digestion fingerprints, as shown in Table 2, the DNA fragment of the wild type will produce a peak of 3738.35 by enzyme digestion, and no It has peaks such as 1577.98, 2196.38, 2485.56, 3778.38 and 3794.37, while the mutant DNA fragment is just the opposite. After digestion and mass spectrometry, one or several of the peaks such as 1577.98, 2196.38, 2485.56, 3778.38 and 3794.37 will be produced, instead of There is a 3738.35 peak. Therefore, in theory, the existence of the wild type can be determined by the presence of the 3738.35 peak, and the presence of the mutant type can be determined by the presence or absence of one or several of the peaks such as...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com