Molecular biological identification method of fritillaria taipaiensis
A technology of molecular biology and Taibai Fritillaria, applied in the field of traditional Chinese medicine identification, can solve the problems of complex splicing and assembly of sequencing data and difficulty in extracting sequencing templates
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
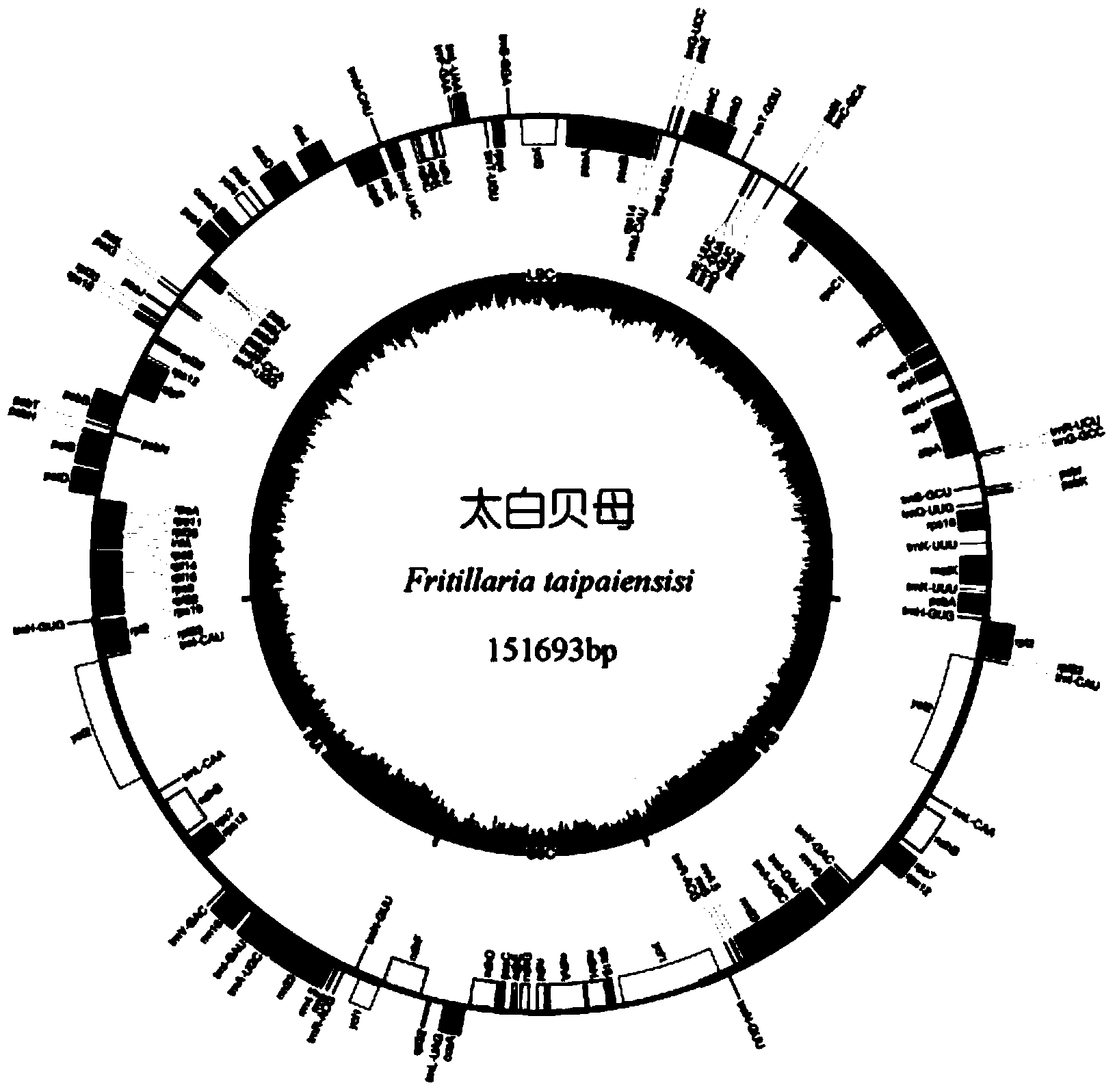
[0068] Example 1 Obtaining of Taibai Fritillaria Chloroplast Genome
[0069] (1) Extraction and purification of chloroplast genomic DNA from Taibai Fritillaria
[0070] Take fresh young leaves of Fritillaria taibai (fresh leaves of Fritillaria taibai were collected from Hongchiba, Wenfengdian, Wuxi County, Chongqing City (latitude: 31.4°, longitude: 109.6°), identified by the Institute of Medicinal Botany, Chinese Academy of Medical Sciences For Taibai Fritillaria.taipaiensis P.Y.Li,), adopt the improved sucrose density gradient centrifugation method to separate, extract and purify the chloroplast genome (cpDNA), the steps are as follows:
[0071] ①Take 100g of fresh young leaves, treat them in the dark at 4°C for 24 hours, wash them, remove the veins, tear them into pieces about 1cm2 in size, add 500mL of buffer A pre-cooled at 4°C, and place them in a homogenizer, so that the leaves are basically submerged. homogenate. Then filter once with 4 layers of gauze and once with ...
Embodiment 2
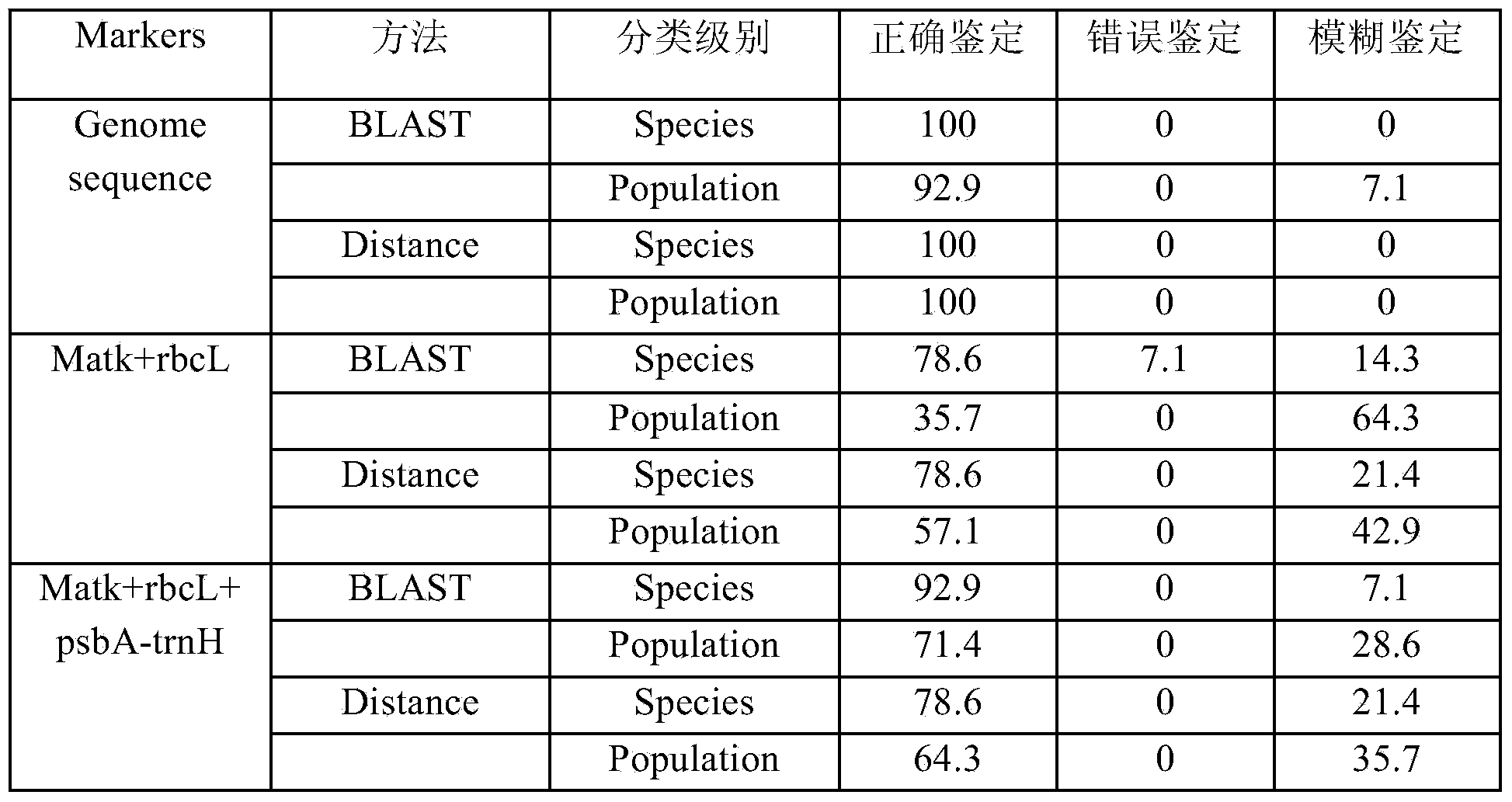
[0093] Example 2 Feasibility of identifying Taibai Fritillaria based on the complete sequence of the chloroplast genome
[0094] (1) Biological barcode identification of Taibai Fritillaria and Chuan Fritillaria based on single-gene sequences
[0095] Genomic DNA of medicinal materials was extracted according to methods known in the prior art, and species identification was performed using chloroplast trnh-psbA sequences. The primers used for the amplification of trnh-psbA region are:
[0096] Upstream primer: 5'-GTTATGCATGACGTAATGCTC-3'
[0097] Downstream primer: 5'-CGCGCATGGTGATTCACAATCC-3'.
[0098] PCR amplification conditions: pre-denaturation at 94°C for 5 min, denaturation at 94°C for 1 min, annealing at 54°C for 1 min, extension at 72°C for 1.5 min, 30 cycles, and finally extension at 72°C for 7 min.
[0099] After sequencing, Fritillaria trnH-psbA sequence:
[0100] ACAATTTCCCTCTAGACCTAGCTGCTATTGAAGTTCCATCTACAAATGGATAAGACTTACATTCTGTCTTAGTGTATACGAATCCTTGATGGAGCAATA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com