DNA sequence for expressing 3 alpha-hydroxysteroid dehydrogenase in Pseudomonas aeruginosa
A technology of hydroxysteroid and DNA sequence, applied in the field of DNA sequence of polypeptide chain and its complementary chain sequence, can solve problems such as high cost, and achieve the effect of high activity and cost reduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] This embodiment is to clone Pseudomonas aeruginosa ( Pseudomonas aeruginosa ) 3α-Hydroxysteroid dehydrogenase 3-HSDA gene fragment, cloned from Pseudomonas aeruginosa ( Pseudomonas aeruginosa ) The 3α-hydroxysteroid dehydrogenase 3-HSDA gene fragment was sequenced and identified to be 1134 bp in length.
Embodiment 2
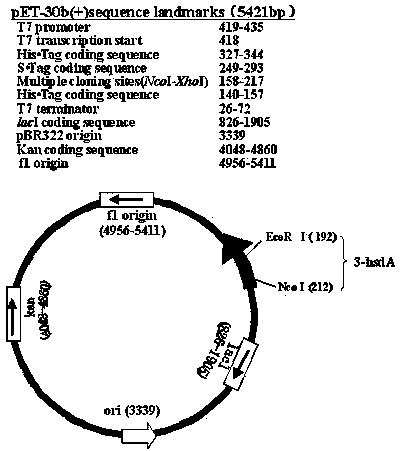
[0055] The present embodiment is Pseudomonas aeruginosa ( Pseudomonas aeruginosa ) The 3α-hydroxysteroid dehydrogenase 3-HSDA gene fragment is recombined with the pET30b(+) expression vector to form the pET30b(+)-hsdA recombinant vector. The steps are as follows:
[0056] A. Pseudomonas aeruginosa ( Pseudomonas aeruginosa ), the size of the original clone of its 3α-hydroxysteroid dehydrogenase gene 3-hsdA is 1134bp, and its polymerase chain reaction (polymerase chain reaction, PCR) system is as follows:
[0057]
[0058] After mixing, carry out PCR reaction according to the following reaction conditions.
[0059]
[0060] Detected by 1% agarose gel electrophoresis, and cut the gel to recover the target fragment.
[0061]
[0062] B. Single digestion of cloning vector pUC19
[0063] The size of the cloning vector pUC19 used for the 3α-hydroxysteroid dehydrogenase gene 3-hsdA is 2686 bp, pUC19 was digested with SmaⅠ, and the enzyme digestion reaction system was as foll...
Embodiment 3
[0081] In this embodiment, after pET30b(+)-hsdA recombinant is transformed into Escherichia coli BL21(DE3), the experimental steps for obtaining BL21(DE3) engineering bacteria of 3α-hydroxysteroid dehydrogenase through screening with kanamycin are as follows:
[0082] Get through kanamycin selection, pET30b (+)-hsdA recombinant vector transforms Escherichia coli BL21 (DE3) the single bacterium colony that forms in the culture medium, inoculate in the 5mlLB culture fluid that contains 50μg / ml kanamycin, 37 Shake culture at ℃ overnight; take 1ml of the culture solution, and extract the plasmid according to the "Plasmid Small Extraction Kit Instructions" of Tiangen Biochemical Technology (Beijing) Co., Ltd.; take 10μl of plasmid DNA, add 4μl of distilled water, 2μl of buffer, 2μl 0.1%BSA and 1μl NcoⅠ, 1μl Eco RⅠ After the enzyme was mixed, it was placed in a water bath at 37°C and incubated for 5 hours; the recombinant DNA and the digested plasmid DNA were electrophoresed at th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com