Vector for knocking out L-lactic dehydrogenase 1 gene and construction method of vector
A technology of lactate dehydrogenase and construction method, which is applied in the field of carrier and construction for knocking out L-lactate dehydrogenase 1 gene, can solve the problems of time-consuming, difficult to succeed and low efficiency of enzymatic digestion and connection, and achieve splicing Effects of improved reaction conditions, reduced use times, and cost and time savings
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
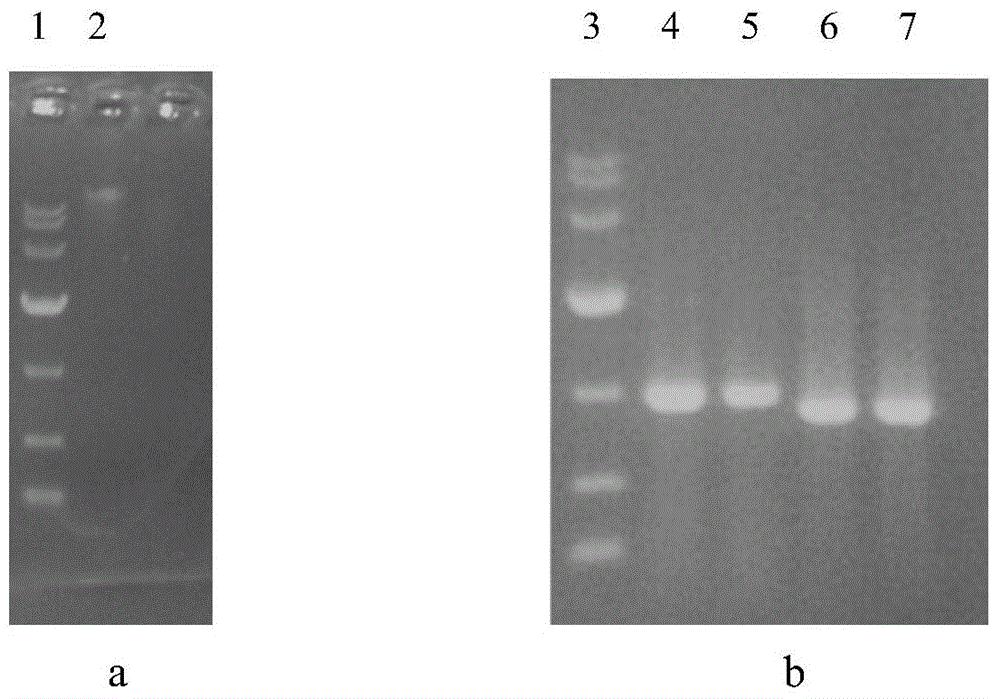
[0031] Example 1 Preparation of Lactobacillus plantarum DM9010 genome and extraction of recombinant plasmid. Including the following steps:
[0032] (1) Centrifuge 1.5ml of the overnight cultured Lactobacillus plantarum solution and collect the precipitate, add 0.6mL of 20mg / ml lysozyme, invert and mix for 5-10 times, then place it at 37°C for 40min;
[0033] (2) Centrifuge at 12,000 rpm for 10 min at room temperature, discard the supernatant carefully.
[0034] (3) Add 150 μL SP Buffer (containing RNase A1) to fully suspend the bacterial pellet.
[0035] (4) Add 20 μL of Lysozyme solution, mix evenly and let stand at room temperature for 5 minutes.
[0036] (5) Add 30 μL of EDTA Buffer, mix evenly and let stand at room temperature for 5 minutes.
[0037] (6) Add 200 μL of Solution A, shake vigorously and keep warm at 65°C for 10 minutes.
[0038] (7) Add 400 μL of Solution B and shake vigorously for 15 seconds.
[0039] (8) Add 650 μL of Solution C and mix evenly by inve...
Embodiment 2
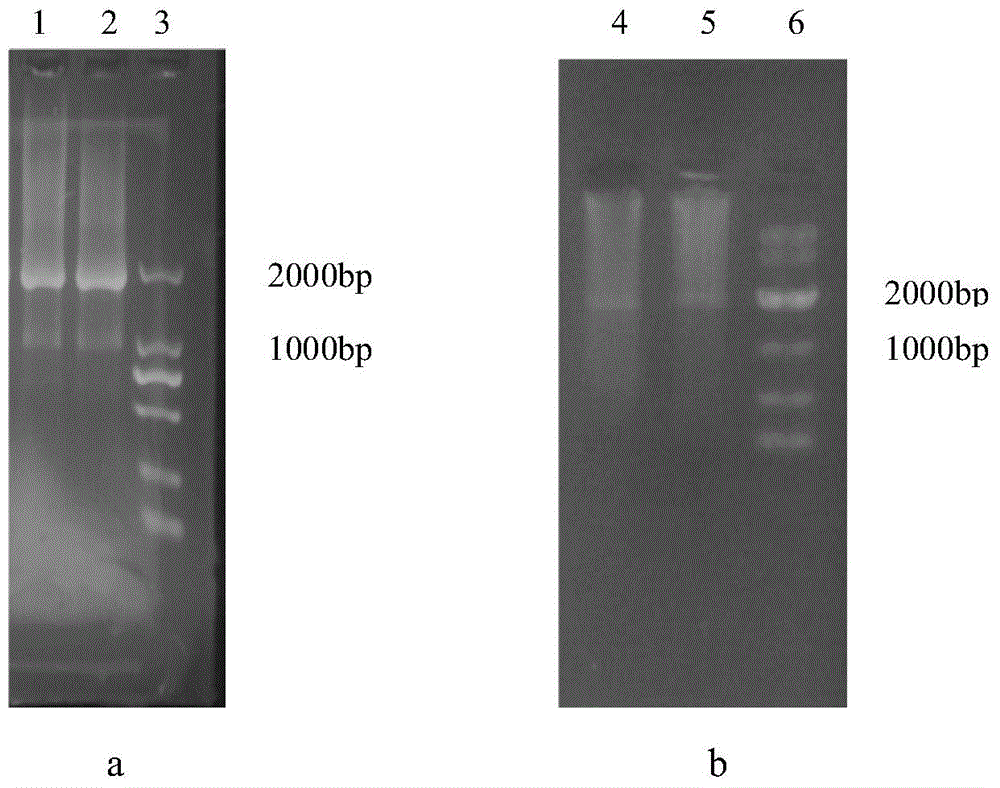
[0052] Example 2 The upstream and downstream fragment primer design and PCR amplification of the Lactobacillus plantarum DM9010L-lactate dehydrogenase 1 gene, the specific operations are as follows:
[0053] (1) According to the reported gene sequence encoding L-lactate dehydrogenase 1, BLAST was performed on Lactobacillus plantarum WCFS1, ST-Ⅲ, JDM1, ZJ316, P8, and 16 whose genomes had been sequenced in NCBI to find L-lactate The location of the gene encoding dehydrogenase 1 and its upstream and downstream DNA sequence of about 1000 bp, then determine the conserved sequence to design primers and introduce restriction sites and 15 bp overlapping fragments.
[0054] (2) PCR amplifies upstream DNA fragment 978bp, downstream fragment 870bp, determines the reaction system of 50 μ l as follows: Lactobacillus plantarum DNA template 5 μ l, 5×PCR PrimerSTAR, Buffer (Mg 2+ plus) 10 μl, dNTP Mixture 4 μl, each primer 1 μl, PrimerSTAR HS DNA Polymerase 0.5 μl, add water to 50 μl. The re...
Embodiment 3
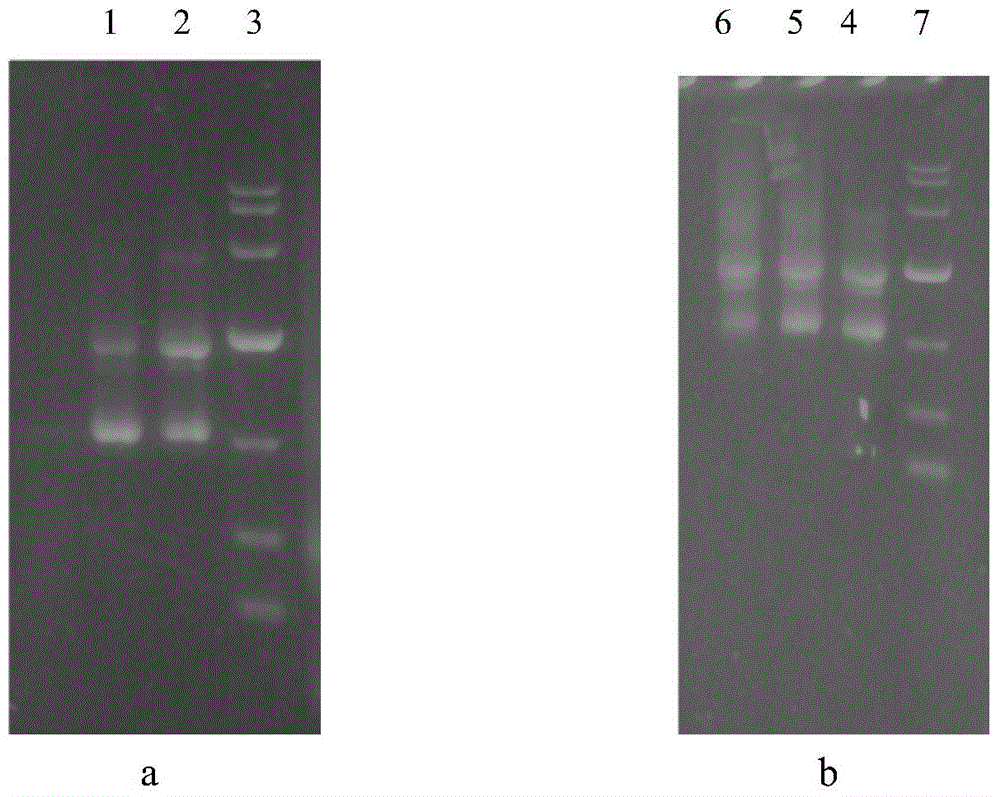
[0055] Example 3 Condition optimization of overlap extension PCR ligation of upstream and downstream DNA fragments. The specific operation is as follows:
[0056] (1) Step-by-step overlap extension PCR reaction process is divided into three steps: (a) Use high-fidelity DNA polymerase to amplify the upstream and downstream fragments respectively, and cut the gel for recovery. (b) Overlap extension of the upstream and downstream DNA fragments, high temperature denaturation during the reaction process to make the upstream and downstream DNA fragments into single strands, and then annealing to pair the overlapping regions (30bp) of the upstream and downstream DNA fragments, and extend each other as templates. The reaction system is as follows: 5×PCR PrimerSTAR, Buffer (Mg 2+ plus) 10 μl, dNTP Mixture 4 μl, upstream DNA fragment 2 μl (about 20 ng), downstream DNA fragment 2 μl (about 20 ng), PrimerSTAR HS DNA Polymerase 0.5 μl, add water to 50 μl. Extension reaction conditions: 9...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com