Application of c-type lectin-3 (cgclec-3) recombinant protein of long oyster
A technology of recombinant protein and oyster growth, applied in the field of molecular biology, can solve the problems of loss, large-scale death economy of cultured oysters, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] In Vitro Prokaryotic Recombinant Expression of C-type Lectin-3 (CgCLec-3) Gene Coding Region in Oyster oyster
[0019] 1. Construction of recombinant vector
[0020] The present invention uses the pET-32a prokaryotic expression vector as the recombinant vector template. By PCR technology, gene-specific primers with specific restriction sites of BamHI and HindIII were added at the 5' ends respectively:
[0021] P1 (5'-CGCGGATCCATGGGTGCCAACTCCATTG-3'):
[0022] P2(5'-CCCAAGCTT TTATTGCGTGTTCCTCGC-3'),
[0023] The reaction conditions were as follows: 94°C pre-denaturation for 5 minutes, followed by the following cycles: 94°C denaturation for 30 seconds, 55°C annealing for 30 seconds, 72°C extension for 45 seconds, a total of 35 cycles, and finally 72°C extension for 10 minutes. The amplified fragment was purified and recovered, and connected to the pMD19-T vector. After transformation, positive clones were screened, and plasmids were extracted; plasmids were digested w...
Embodiment 2
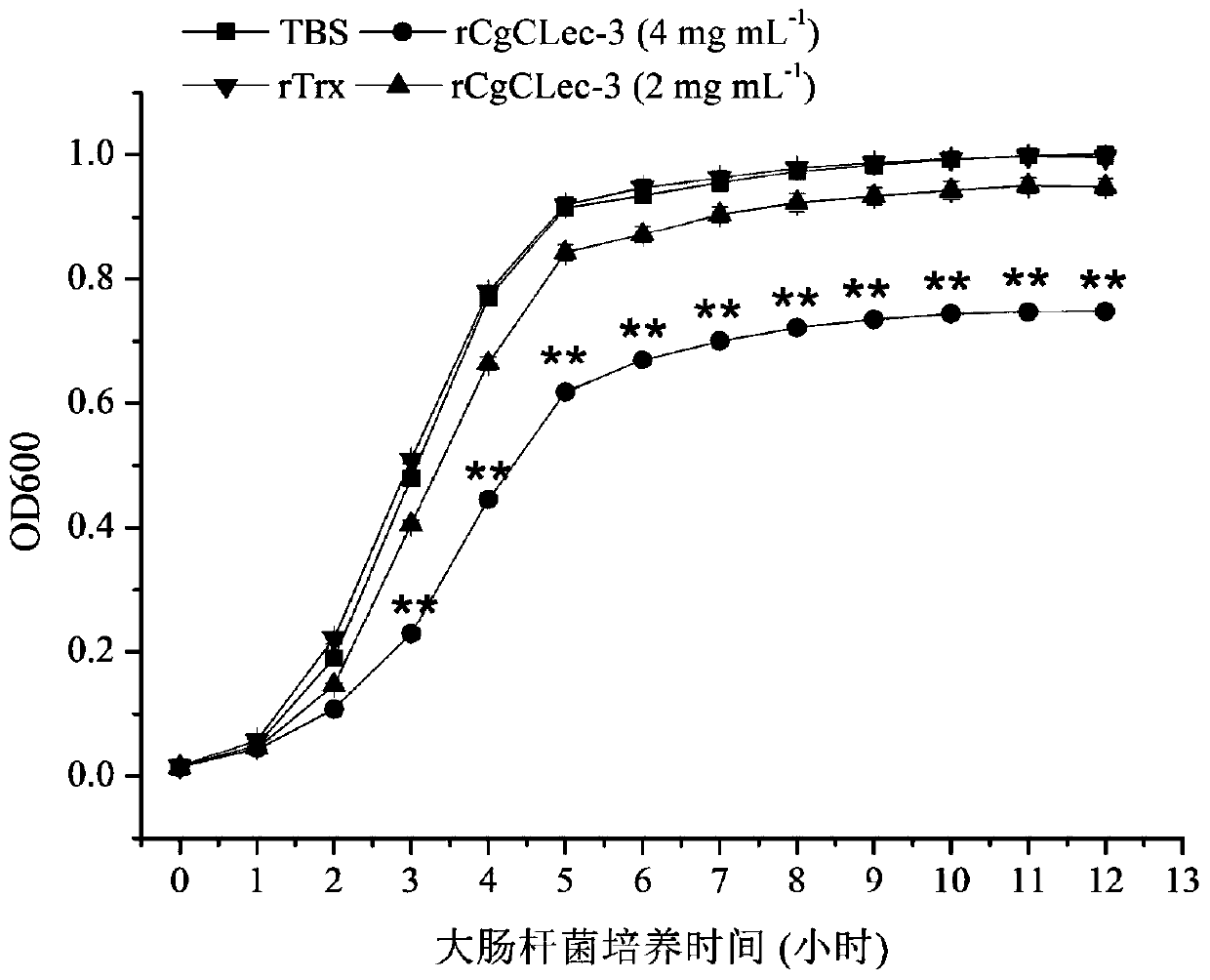
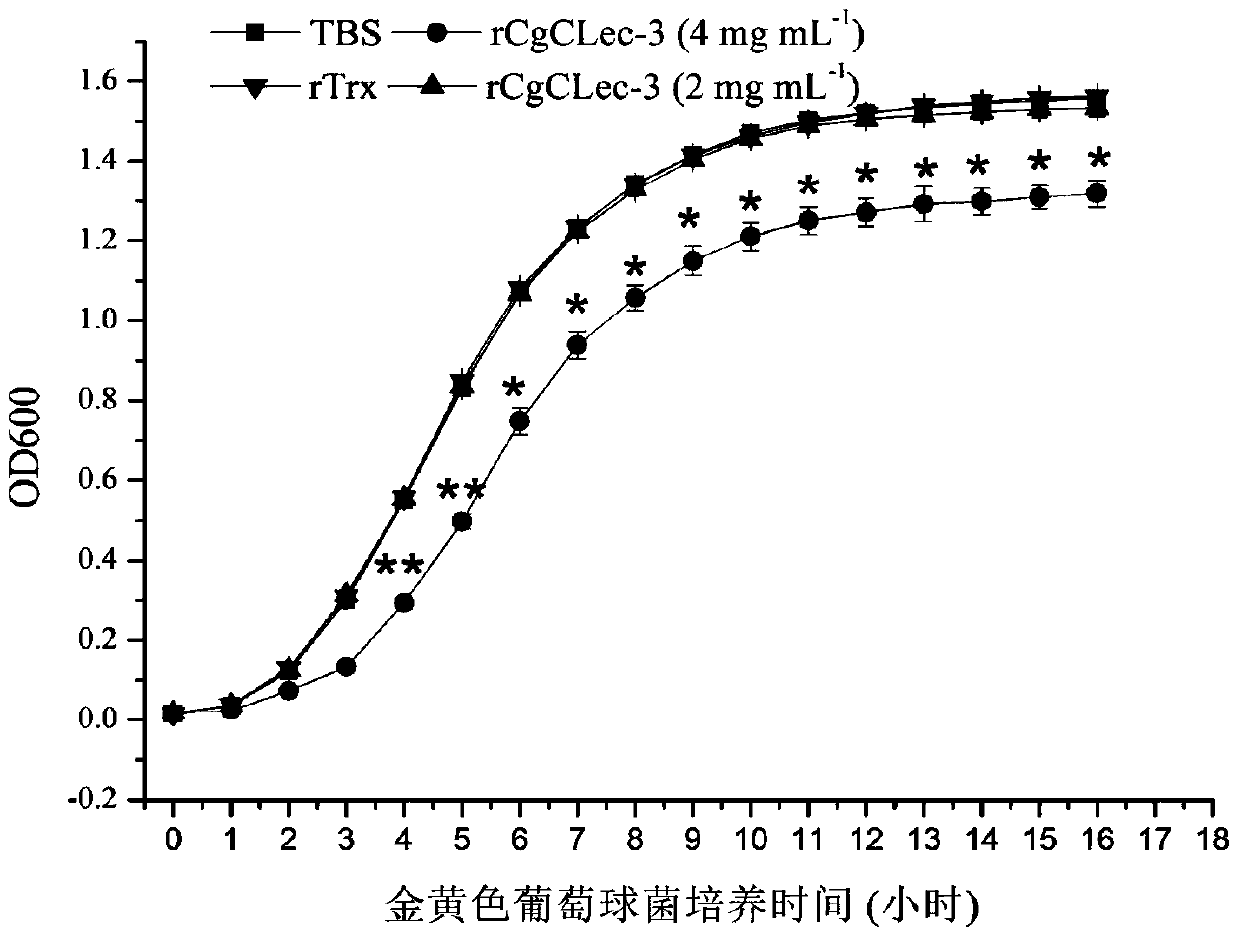
[0037] Detection of Bacteriostatic Activity of CgCLec-3 Recombinant Protein from Oyster oyster
[0038] Escherichia coli and Staphylococcus aureus were cultured in LB medium at 37°C and 220rpm until logarithmic growth phase, and the obtained bacteria were collected by centrifugation and washed with TBS (50mM Tris-Hcl, 100mM NaCl, pH=8.0) and weighed. Suspended to a concentration of 10 4 CFU.
[0039] 50 μL of the above-mentioned or obtained long oyster recombinant protein CgCLec-3 (2 mg mL -1 or 4mg mL -1 ) with an equal volume of Escherichia coli or Staphylococcus aureus suspension in CaCl 2 Incubate at room temperature for 2 h at a final concentration of 10 mM. Take 20 μL of the above mixture in a flat-bottomed 96-well plate (Costar, Fisher), add 200 μL LB liquid medium to each well, shake and culture in a microplate reader at 37°C, measure and record the OD600 value of each well every 1 h. Another rTrx protein control group and TBS blank control group were set up. Thr...
Embodiment 3
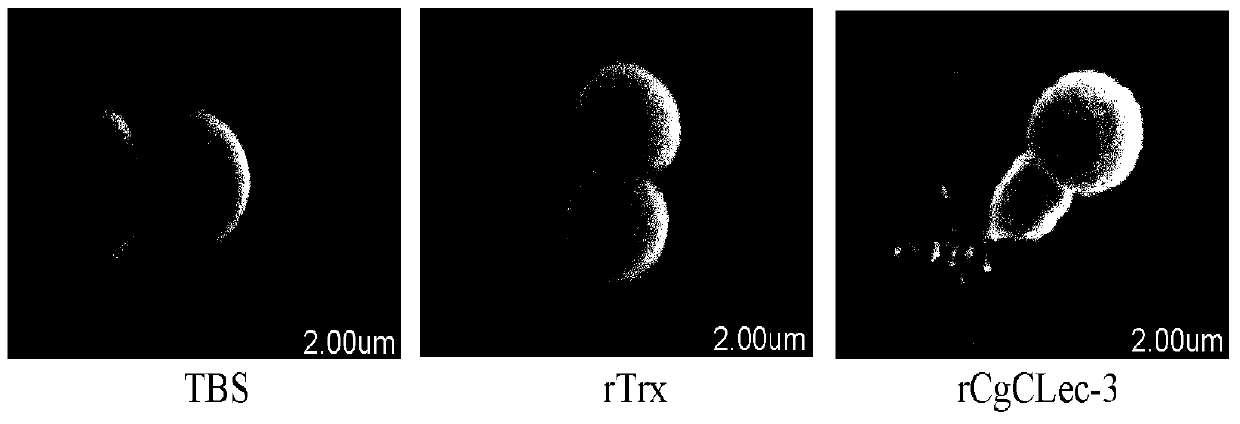
[0041] Determination of the destructive effect of recombinant protein CgCLec-3 on Staphylococcus aureus cells
[0042] After the Staphylococcus aureus cultured to the logarithmic growth phase in the above-mentioned embodiment 2 was collected by centrifugation, it was washed and resuspended with TBS (10 4 CFU). At the same time, 50 μL of the long oyster recombinant protein CgCLec-3 (4mg mL) obtained above -1 ) with an equal volume of Staphylococcus aureus suspension in CaCl 2 Incubate at room temperature for 2 h at a final concentration of 10 mM. Set up TBS blank control and rTrx negative control. The incubated bacteria were thoroughly washed and collected by centrifugation. After fixing with 2.5% glutaraldehyde at 4°C for 24h, the bacteria were thoroughly washed with TBS, resuspended in TBS, and dropped on 24×24mm coverslips. After fixing in 2.5% glutaraldehyde for 1 h, the coverslips were rinsed in TBS for 10 min and then washed with amyl acetate for 30 min. After gradi...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap