Delivery, engineering and optimization of systems, methods and compositions for sequence manipulation and therapeutic applications
A composition, engineering technology, applied in the direction of biochemical equipment and methods, other methods of inserting foreign genetic materials, drug combinations, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0751] Example 1: CRISPR complex activity in the nucleus of eukaryotic cells
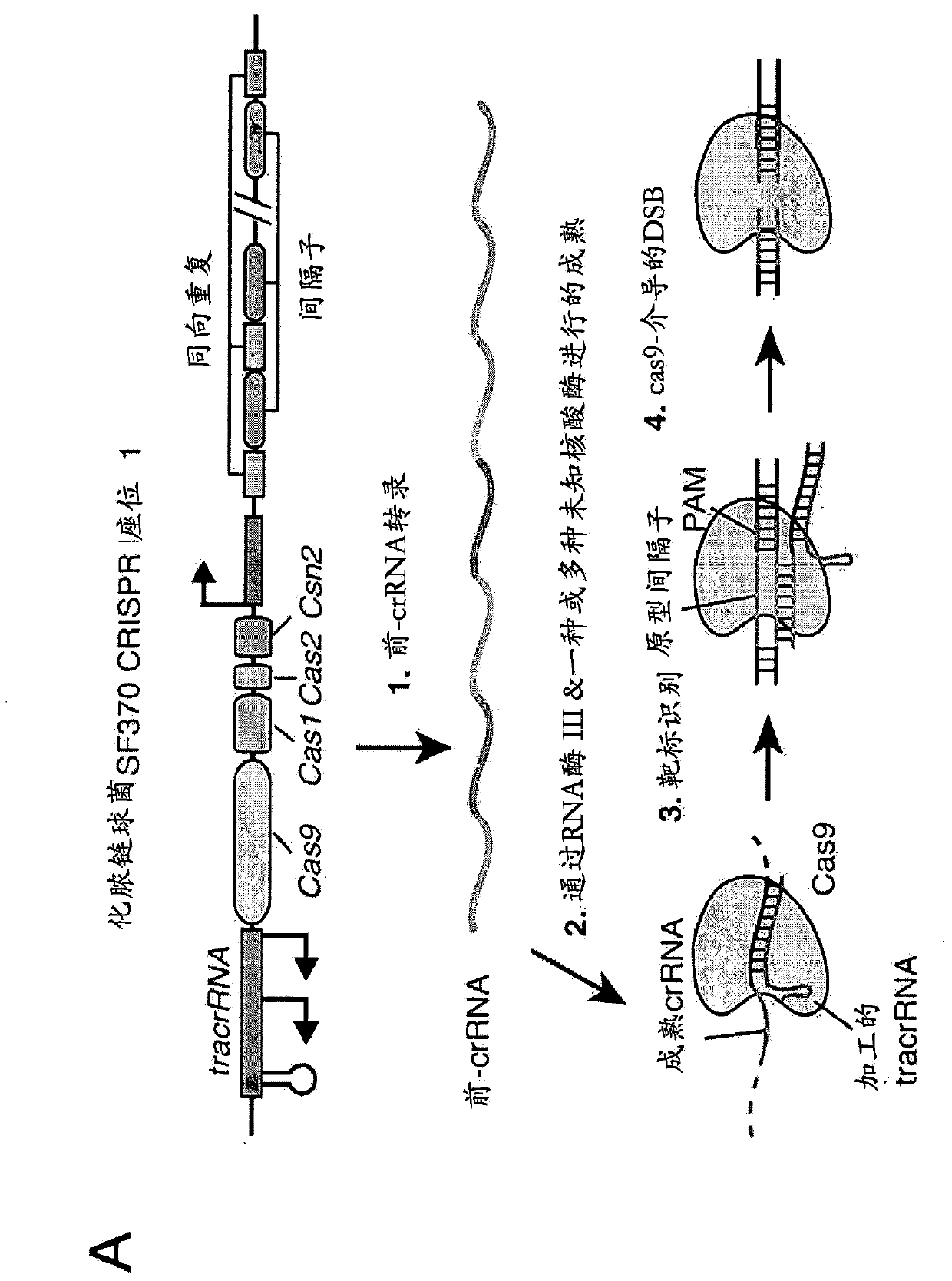
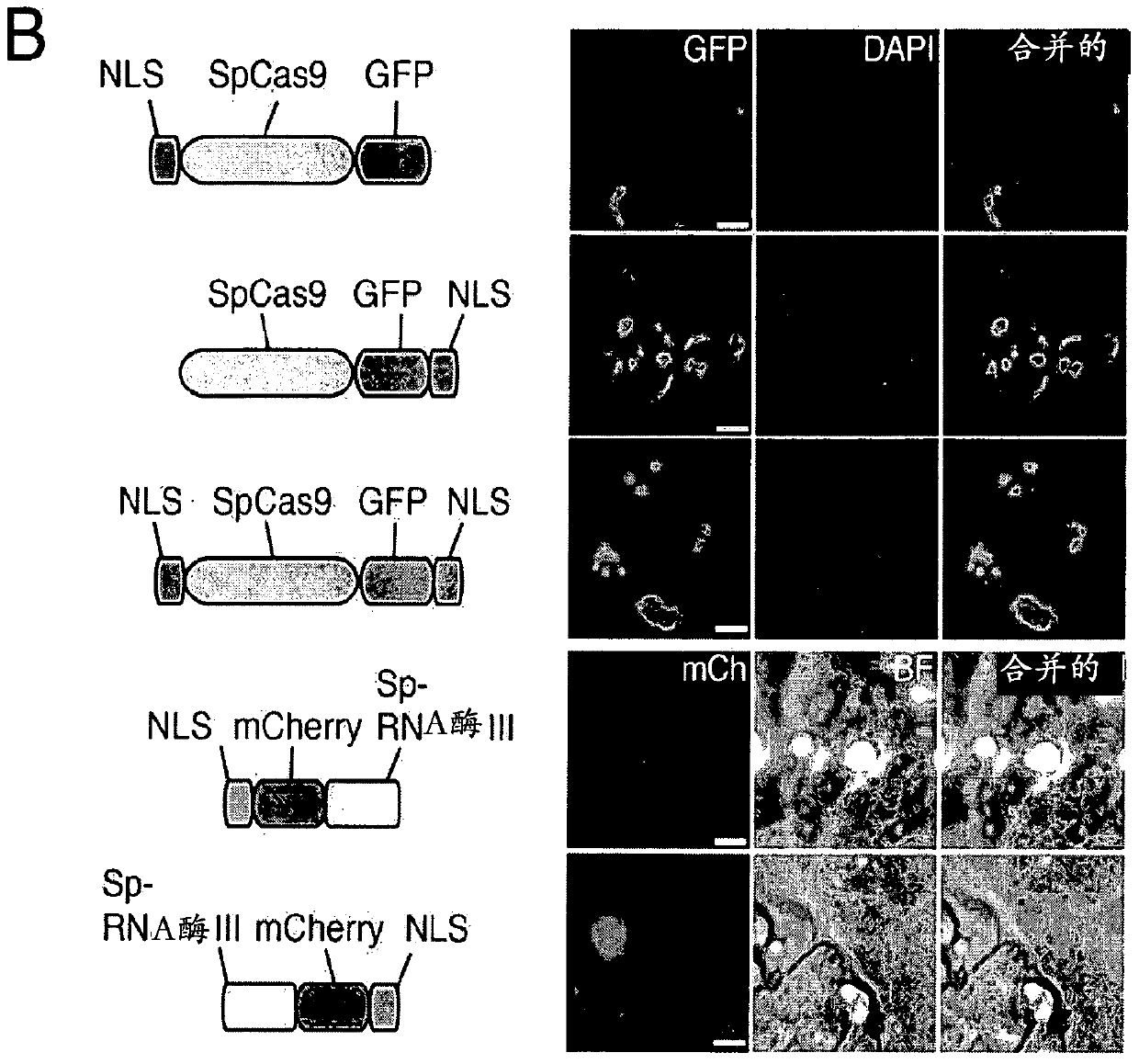
[0752] An exemplary type II CRISPR system is the type II CRISPR locus from S. pyogenes SF370, which contains a cluster of four genes Cas9, Cas1, Cas2, and Csn1 and two non-coding RNA elements tracrRNA and a short sequence composed of non-repetitive sequences. A characteristic array of repeated sequences (direct repeats) spaced apart by segments (spacers, about 30 bp each). In this system, targeted DNA double-strand breaks (DSBs) are generated in four sequential steps ( Figure 2A ). In the first step, two non-coding RNAs, the pre-crRNA array and the tracrRNA, are transcribed from the CRISPR locus. In the second step, the tracrRNA is hybridized to the direct repeat of the pre-crRNA, which is then processed into a mature crRNA containing a separate spacer sequence. In the third step, the mature crRNA:tracrRNA complex guides Cas9 to a DNA target consisting of the protospacer and the corresponding PA...
example 2
[0783] Example 2: CRISPR System Modifications and Alternatives
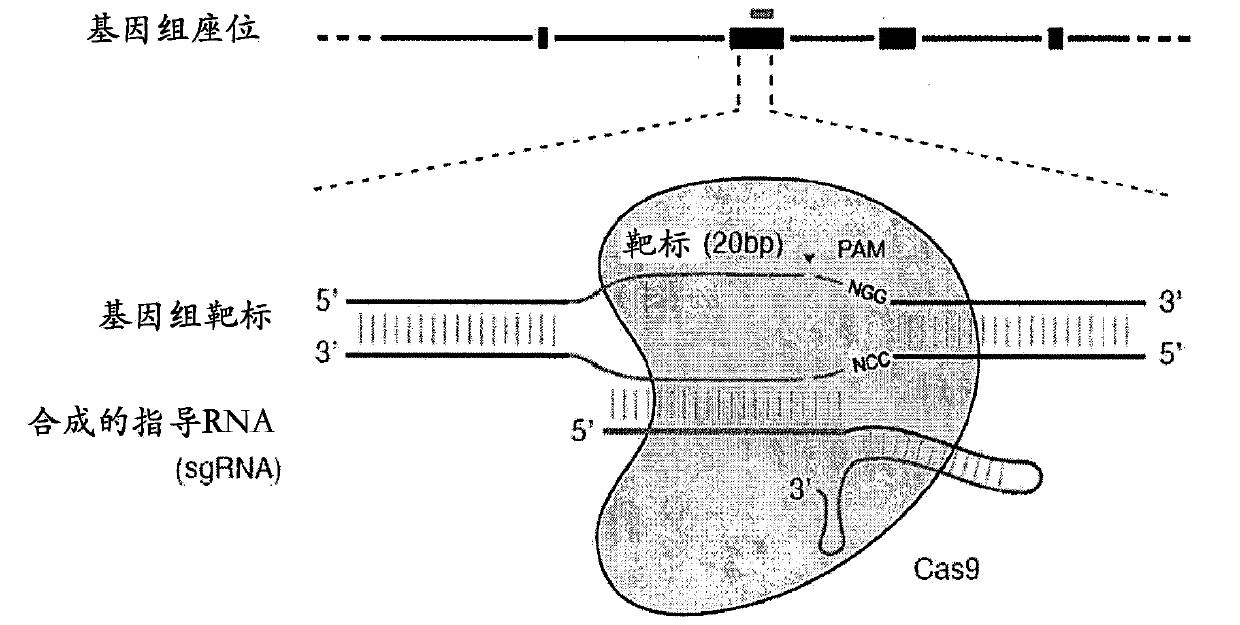
[0784] The ability to program sequence-specific DNA cleavage using RNA defines a new class of genome engineering tools for a variety of research and industrial applications. Several aspects of the CRISPR system can be further improved to increase the efficiency and versatility of CRISPR targeting. Optimal Cas9 activity can be dependent on free Mg present in mammalian nuclei 2+ High levels of free Mg 2+Availability of NGG motifs (see e.g. Jinek et al., 2012, Science, 337:816), and a preference for NGG motifs located just downstream of protospacers limits targeting to the human genome average capacity per 12-bp in (Fig. 9, both positive and negative strands of human chromosomal sequences were evaluated). Some of these constraints can be overcome by exploring the diversity of CRISPR loci across microbial metagenomes (see eg Makarova et al., 2011, Nat Rev Microbiol, 9:467). Other CRISPR loci can be grafted into t...
example 3
[0785] Example 3: Sample target sequence selection algorithm
[0786] A software program is designed to identify candidate CRISPR target sequences on both strands of the input DNA sequence based on the desired guide sequence length and CRISPR motif sequence (PAM) for the specified CRISPR enzyme. For example, the target site of Cas9 from S. pyogenes with the PAM sequence NGG can be identified by searching for 5'-Nx-NGG-3' on both the input sequence and the reverse complement of the input sequence. Likewise, a target site for Cas9 of S. pyogenes CRISPR1 with the PAM sequence NNAGAAW can be identified by searching for 5'-Nx-NNAGAAW-3' both on the input sequence and the reverse complement of the input sequence. Likewise, the target site of Cas9 of S. pyogenes CRISPR3 with the PAM sequence NGGNG can be identified by searching for 5'-Nx-NGGNG-3' on both the input sequence and the reverse complement of the input sequence. Can be fixed by program or specified by userN x The value "x...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle size | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com