Prediction method of protein three-dimensional structure based on bsa-ts algorithm
A technology of three-dimensional structure and prediction method, which is applied in the field of bioinformatics protein structure prediction to improve the global search ability and algorithm accuracy, the algorithm framework is simple and efficient, and the diversity is guaranteed.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0058] The embodiments of the present invention are implemented on the premise of the technical solutions of the present invention, and detailed implementation methods and specific operation processes are given, but the protection scope of the present invention is not limited to the following embodiments.
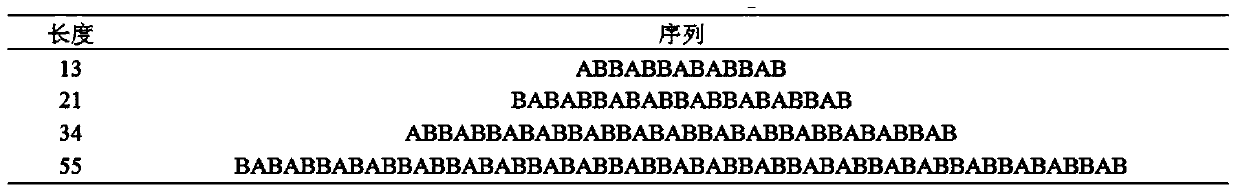
[0059] Because there are many kinds of protein sequences, we use image 3 Take the Fibonacci sequence of length 13 as an example.
[0060] Step 1: Divide the protein sequence into hydrophobic amino acids (H) and hydrophilic amino acids (P), and abstract the protein sequence into a sequence of numbers for computer operations, where the hydrophobic amino acid corresponds to the number 1, and the hydrophilic amino acid corresponds to the number - 1. So a sequence of length 13 is represented as [1 -1 -1 1 -1 -1 1 -1 1 -1 -1 1 -1].
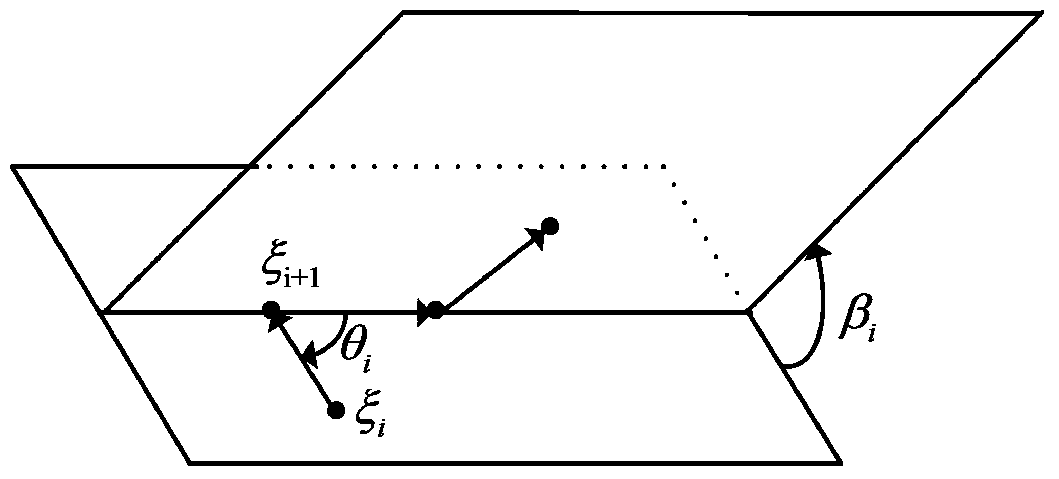
[0061] Step 2: Randomize the population individuals according to the sequence length: when the sequence length is n, dim (problem dimension) is 2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com