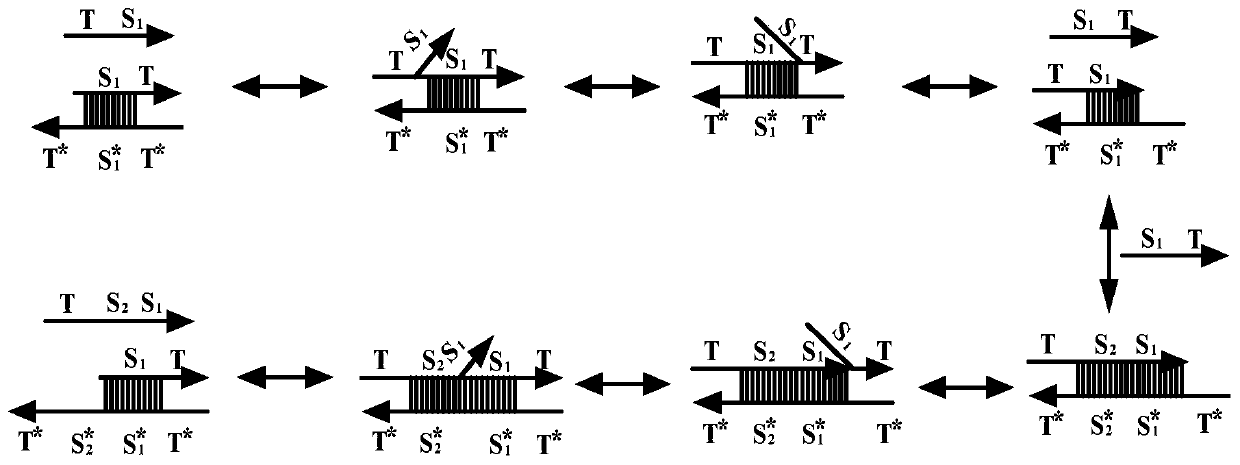
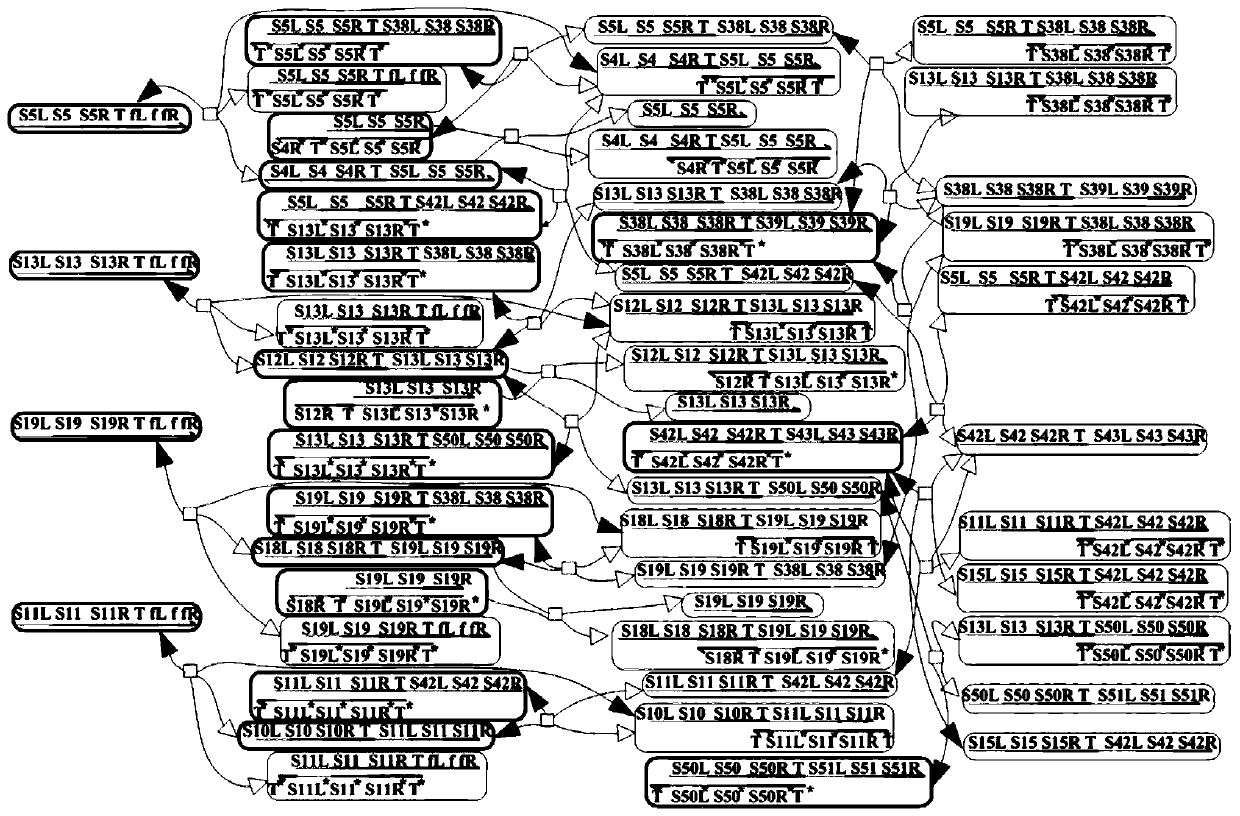
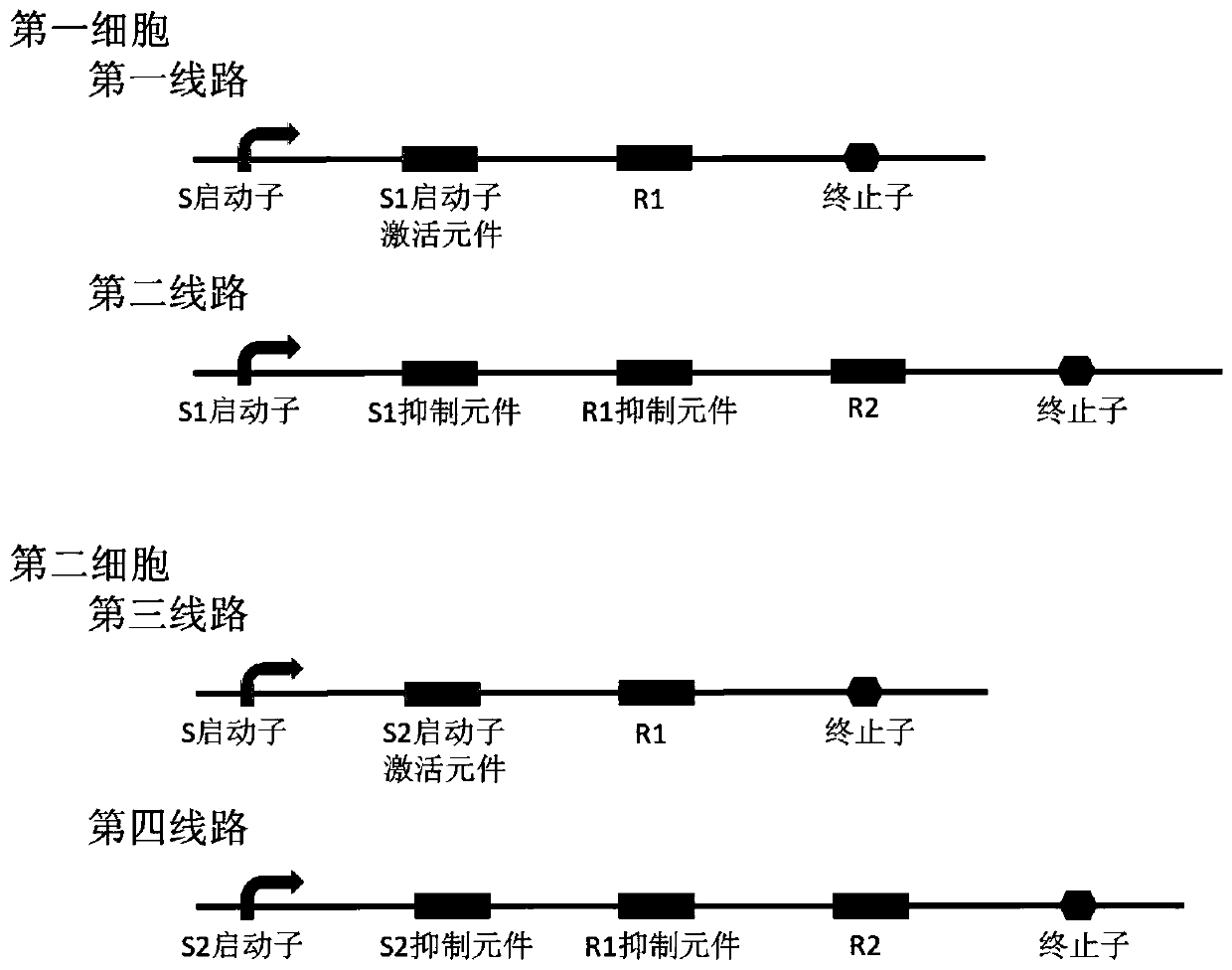
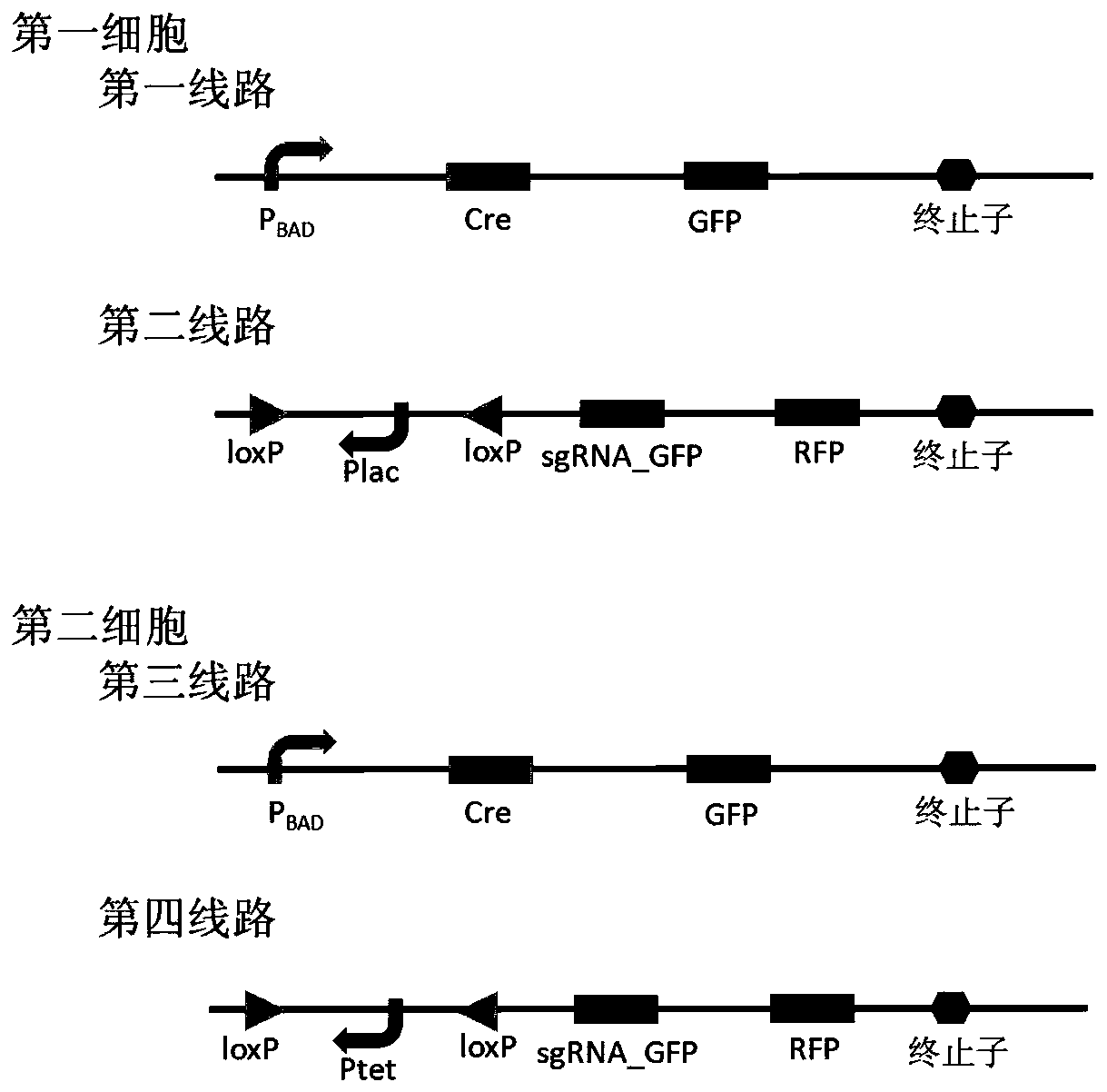
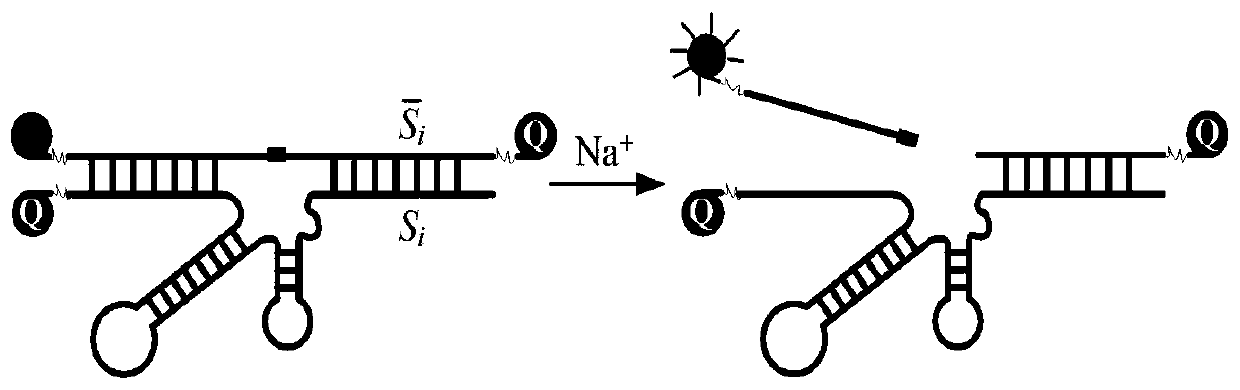
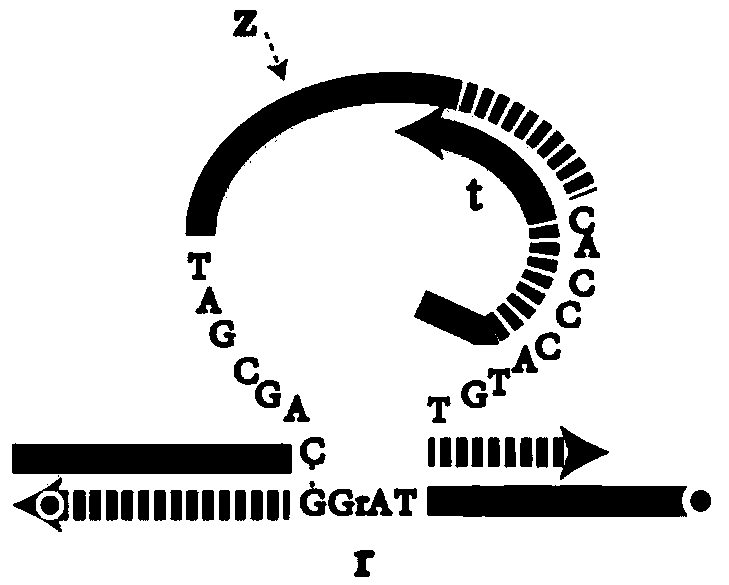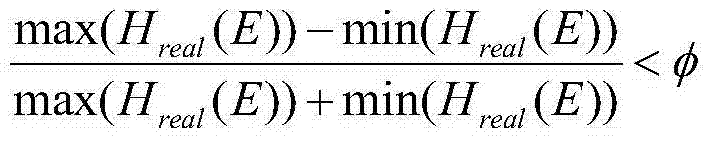Patents
Literature
34 results about "Biocomputer" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Bio computers use systems of biologically derived molecules—such as DNA and proteins—to perform computational calculations involving storing, retrieving, and processing data. The development of biocomputers has been made possible by the expanding new science of nanobiotechnology. The term nanobiotechnology can be defined in multiple ways; in a more general sense, nanobiotechnology can be defined as any type of technology that uses both nano-scale materials (i.e. materials having characteristic dimensions of 1-100 nanometers) and biologically based materials. A more restrictive definition views nanobiotechnology more specifically as the design and engineering of proteins that can then be assembled into larger, functional structures The implementation of nanobiotechnology, as defined in this narrower sense, provides scientists with the ability to engineer biomolecular systems specifically so that they interact in a fashion that can ultimately result in the computational functionality of a computer.
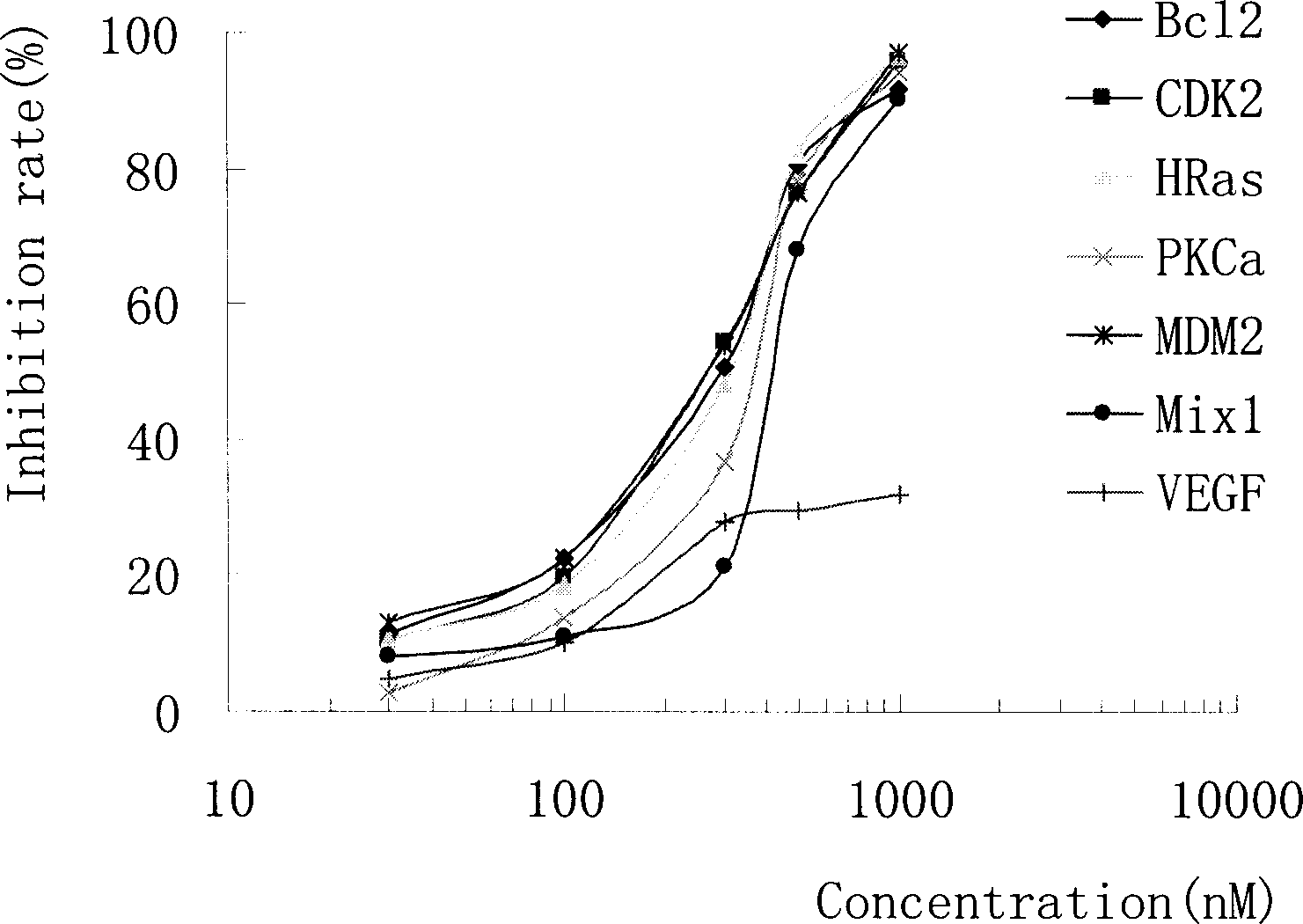
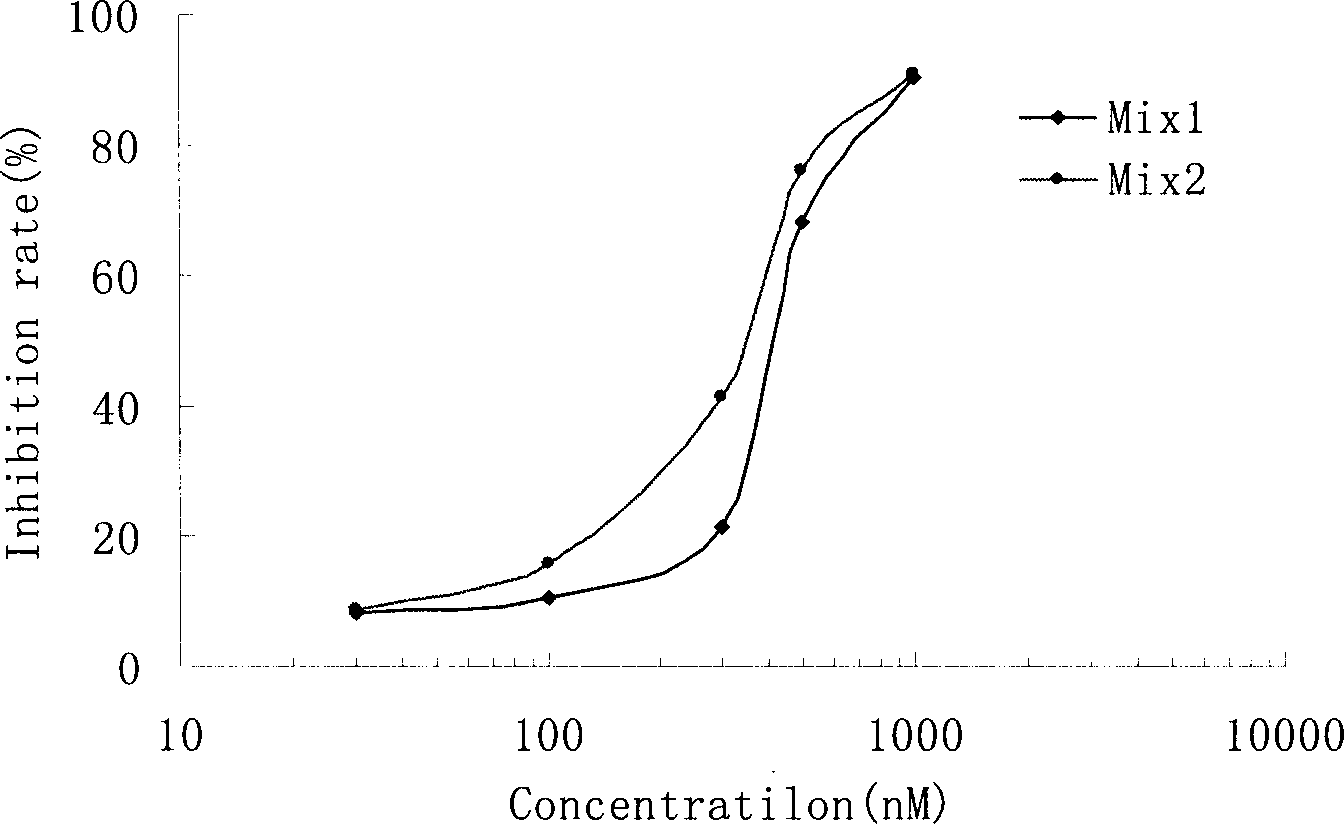
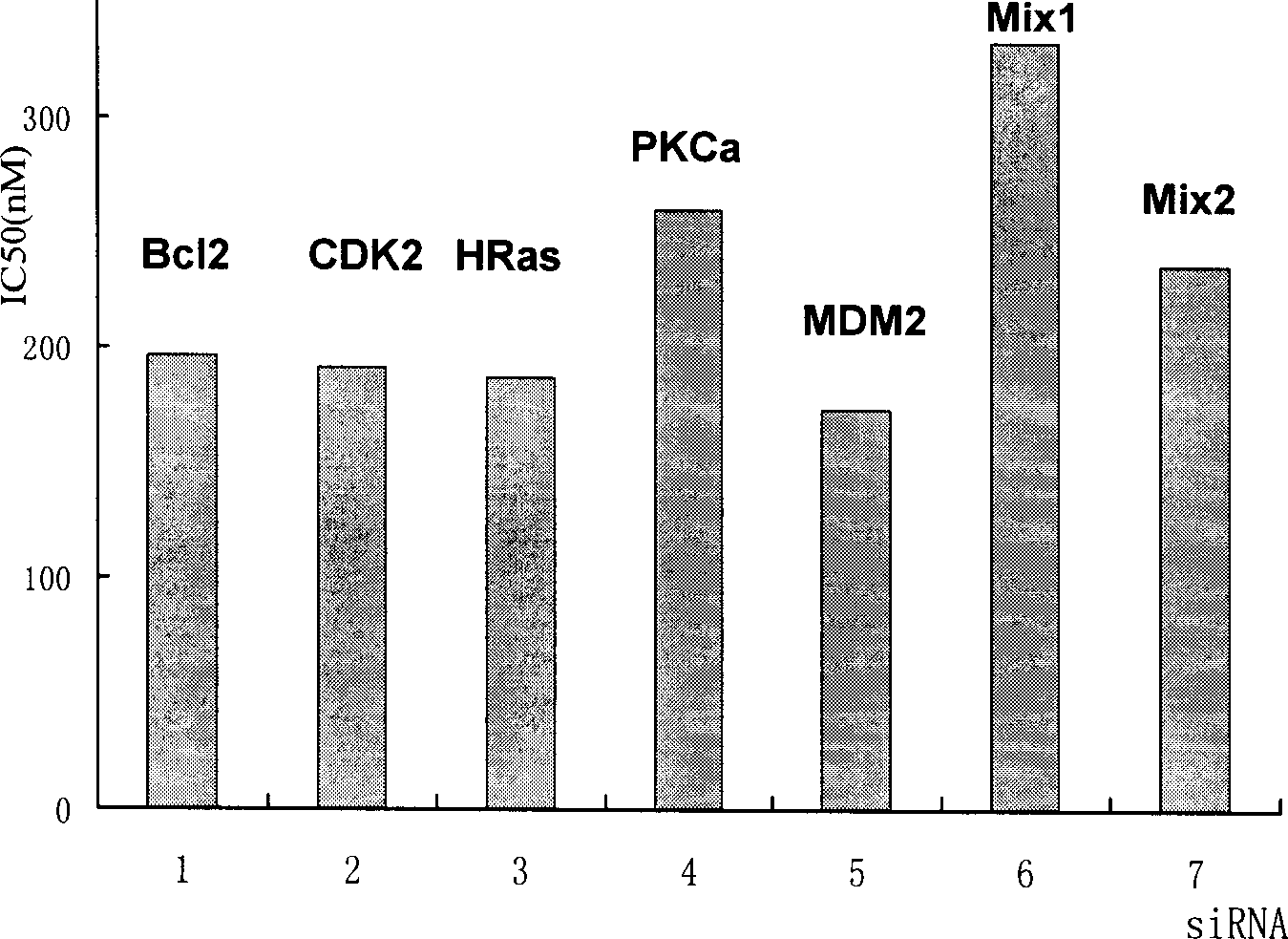
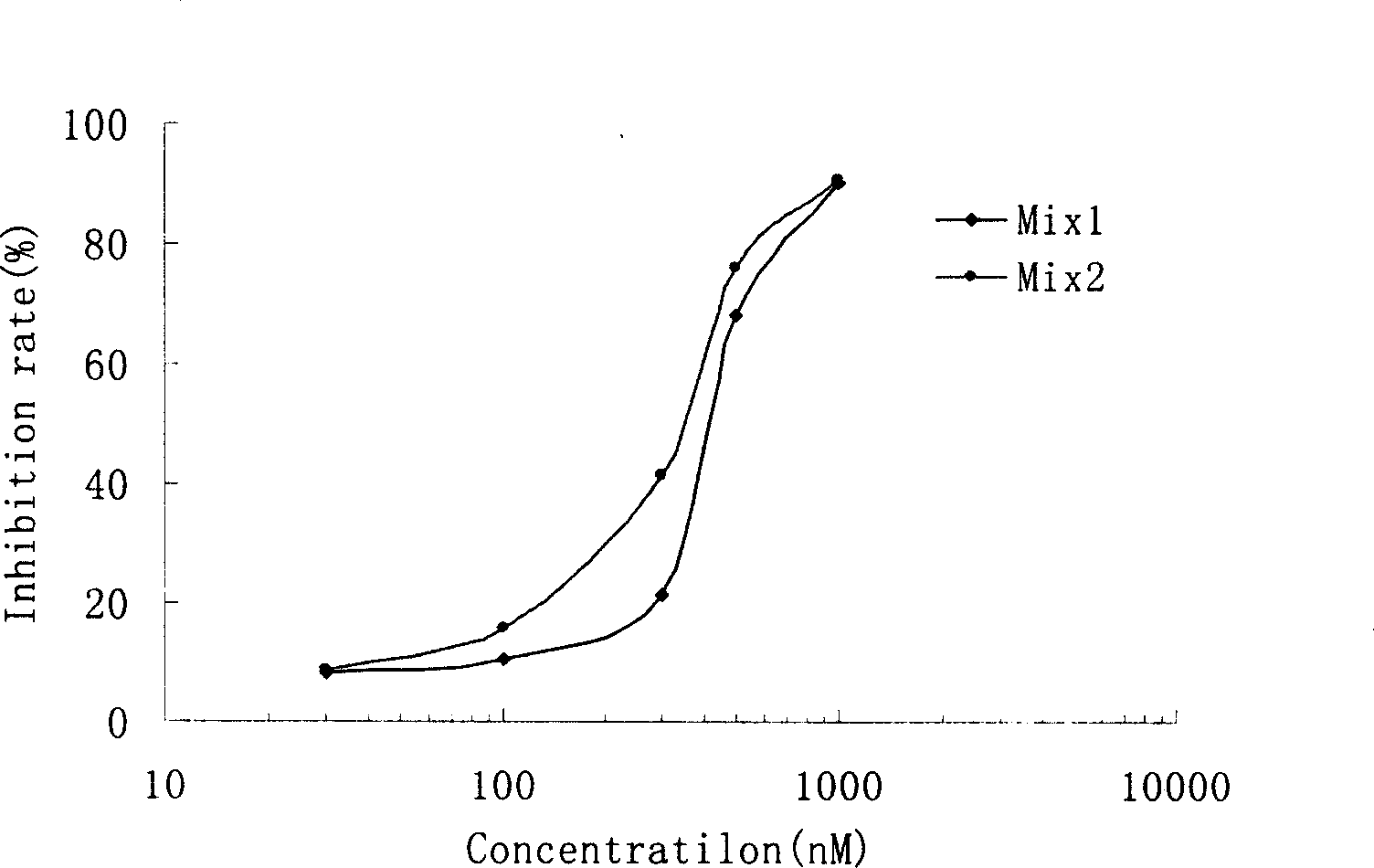
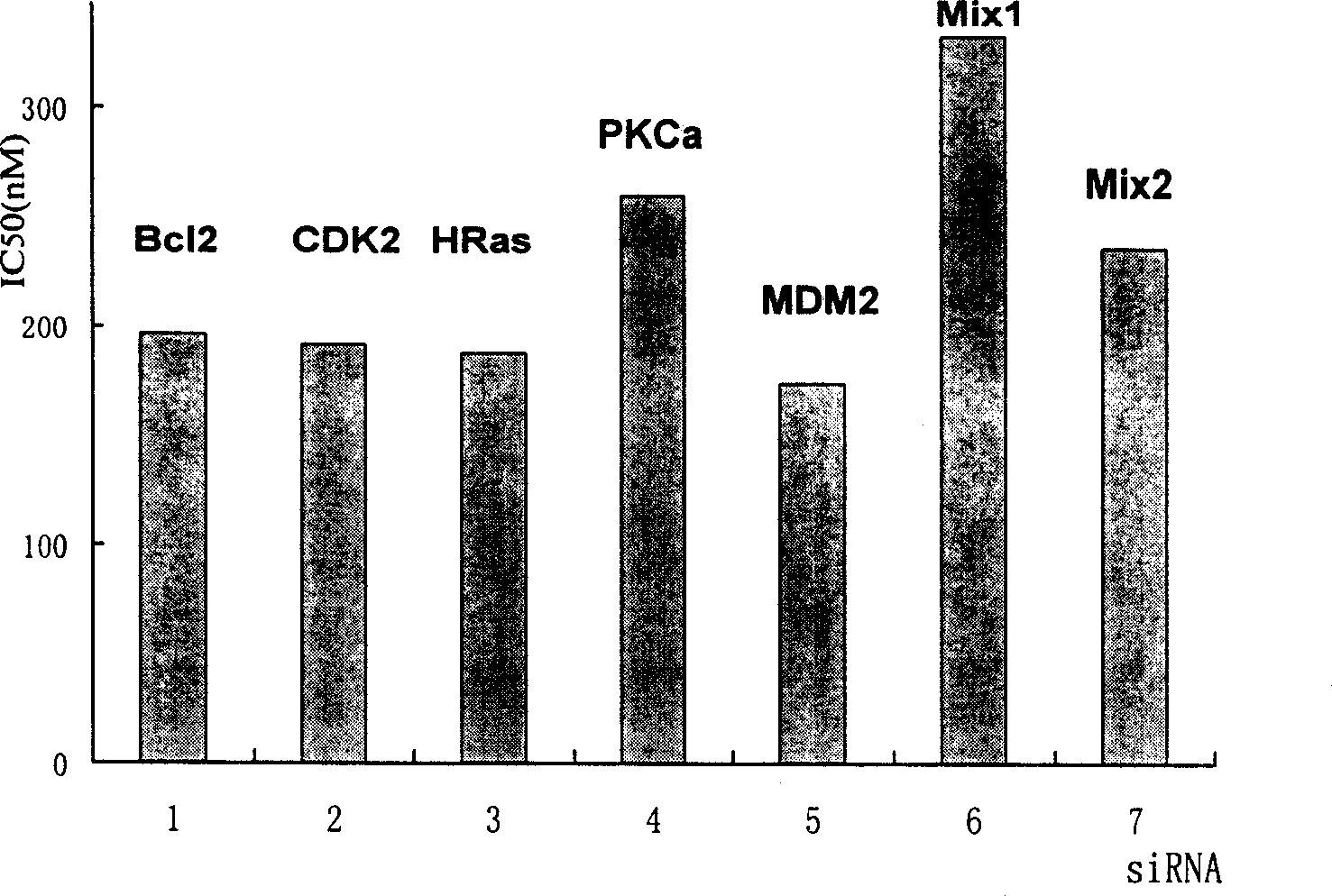
Short interference ribonucleic acid as novel anti-tumor gene therapeutic medicine
InactiveCN1425466AOrganic active ingredientsGenetic material ingredientsHuman tumorComputers technology
By means of biological computer technology, one group of gene sequence relevant to human tumor is obtained from GenBank. Based on RNA interference technology, one group of siRNA capable of inducing RNA interference is designed. Certain amount of siRNA is synthesized chemically and used as novel efficient and specific antitumor medicine. MTT detection shows that siRNA has obvious cytotoxic effect on MCF-7, BEL 7402, KB, HT-29 and other tuomr cells. Western Blotting detection shows that siRNA can lower the expression level of specific protein by 80-90%. Morphological observation shows that siRNA can cause tumor cell produce corresponding morphological change and chromatin concentration. Compared with available technology, siRNA is one antitumor medicine with completely new mechanism, high efficiency and fast speed.
Owner:MEDICINE & BIOENG INST OF CHINESE ACAD OF MEDICAL SCI
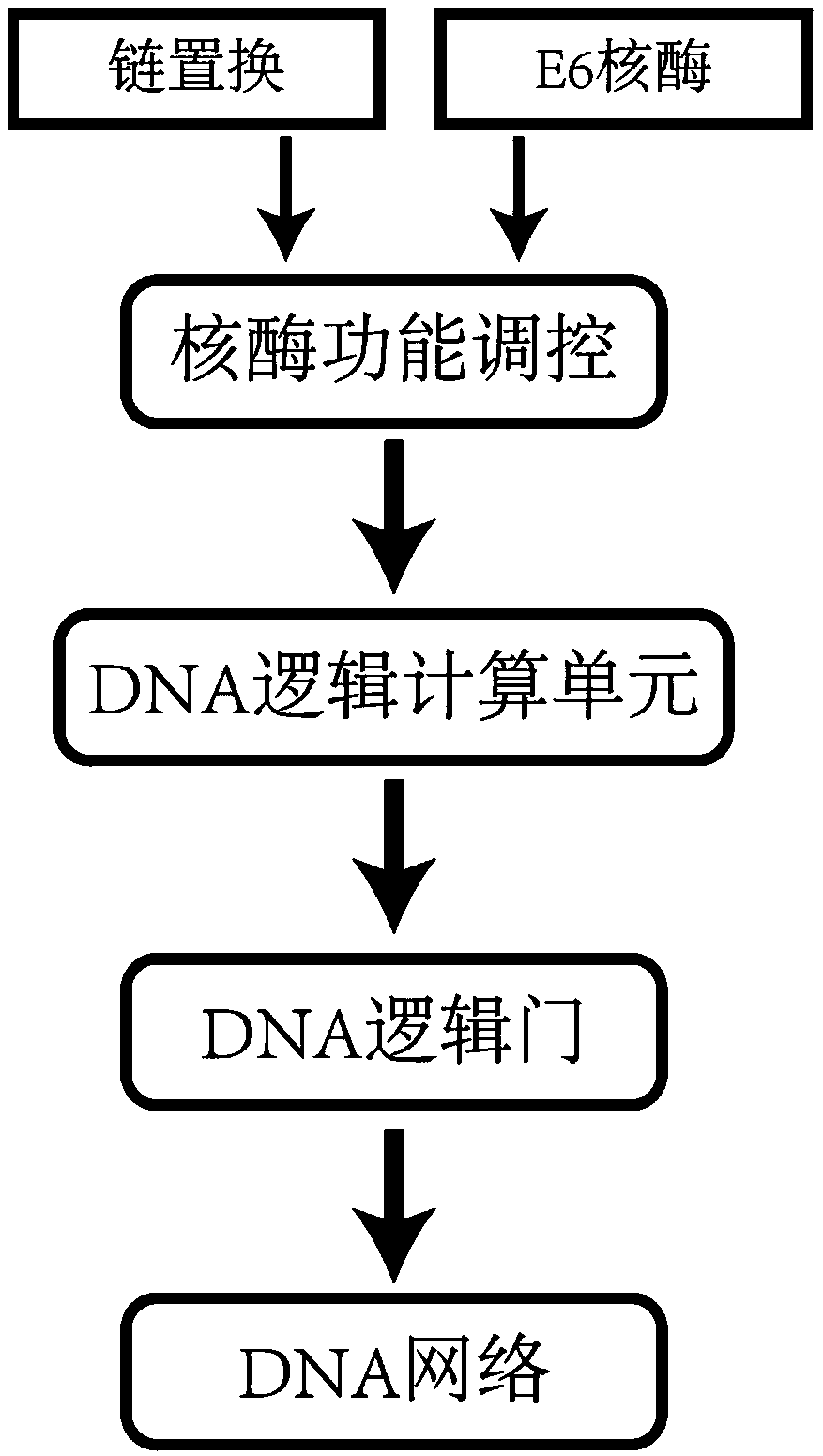
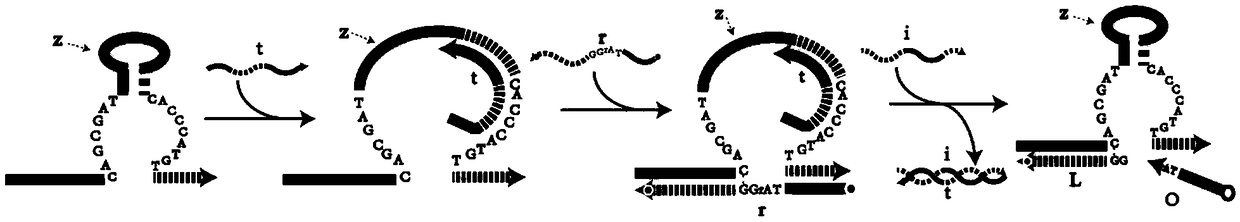
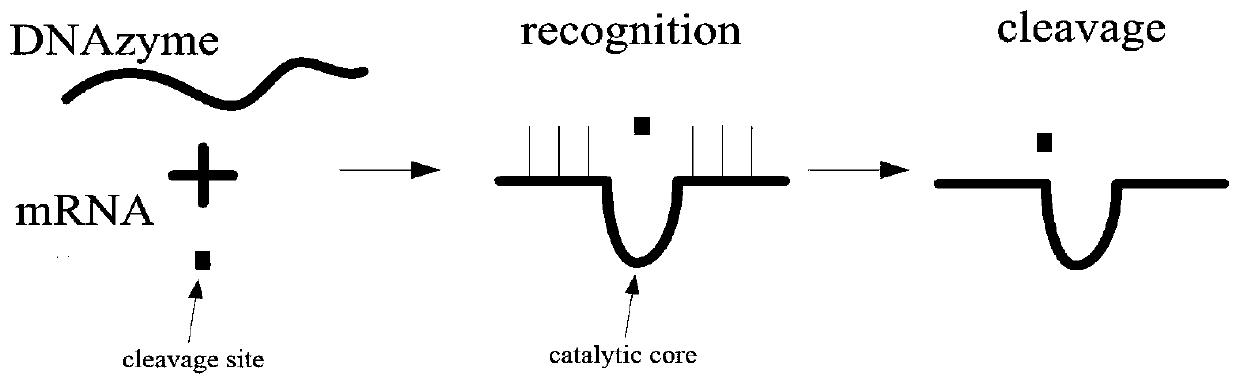
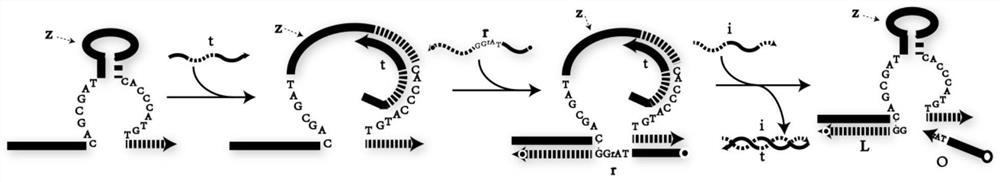
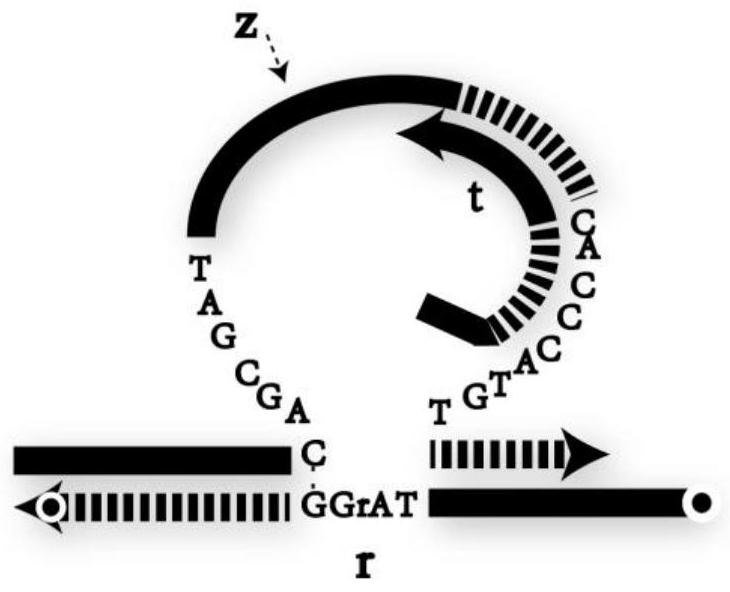
DNA network construction method for regulating and controlling E6 ribozyme function based on chain replacement
ActiveCN108710780ASpecial data processing applicationsMolecular structuresNano structuringLow leakage
The invention relates to a DNA network construction method for regulating and controlling an E6 ribozyme function based on chain replacement. The method comprises the following steps: implementing regulation and control of the E6 ribozyme function through a chain replacement technology according to a clip-shaped two-stage structure and a catalytic core conservative domain sequence of E6 ribozyme;adding an RNA-modified DNA substrate by using a substrate combination arm of the E6 ribozyme, so as to form a branch annular structure and construct a DNA logic calculation unit; constructing a DNA logic gate by transforming the DNA logic calculation unit; and finally connecting the DNA logic gate to form a DNA network. The method cannot destroy the sequence integrity of the E6 ribozyme; and the DNA logic calculation unit has the characteristics of structural stability, customization, modularization, low leakage and interference resistance and can be widely applied to biological calculation and DNA nano structures.
Owner:DALIAN UNIV
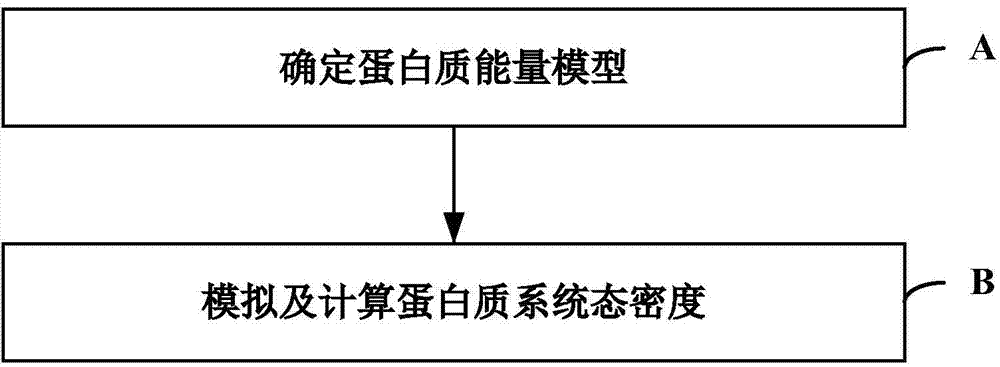
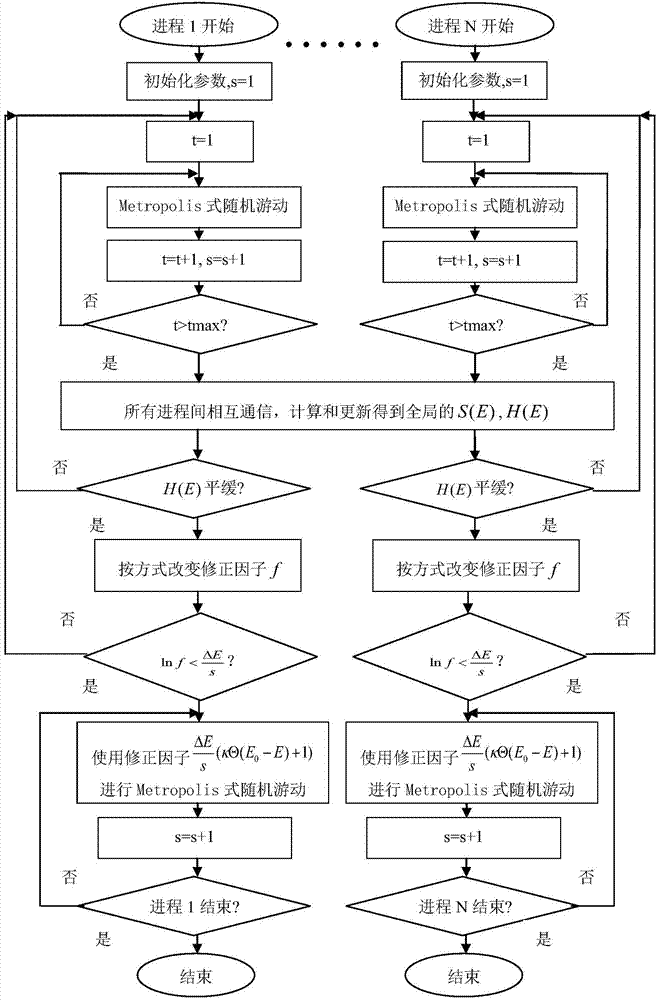
Monte Carlo simulation-based protein thermomechanical-analysis method
ActiveCN103699816AEfficient analysisSpeed up searchSpecial data processing applicationsSimulation basedProtein thermodynamics
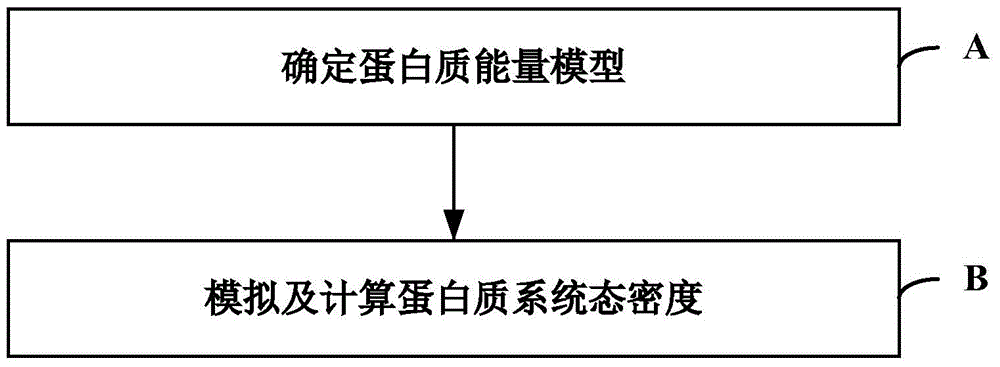
The invention relates to the technical fields of biological computing and Monte Carlo simulation, and particularly discloses a Monte Carlo simulation-based protein thermomechanical-analysis method, which comprises the following steps of A, determining a protein energy model; B, simulating and calculating the state density of a protein system. According to the method, the whole thermomechanical process of protein folding can be efficiently analyzed and researched to further explore and research a protein folding process.
Owner:SHENZHEN INST OF ADVANCED TECH
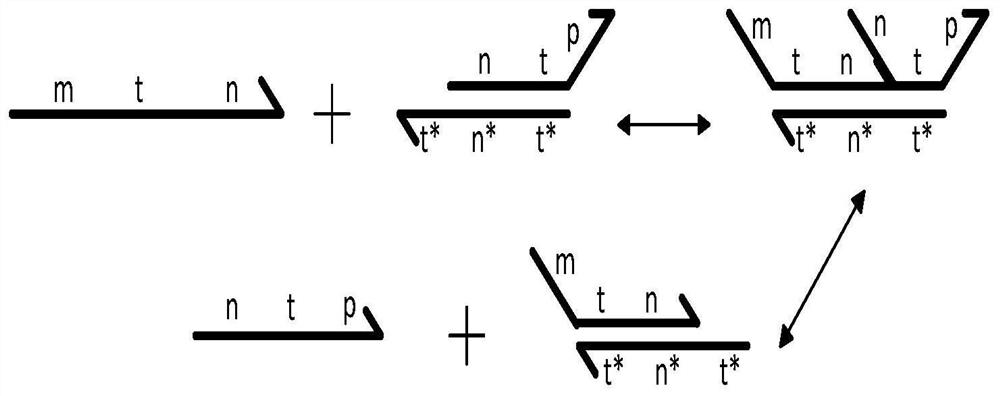
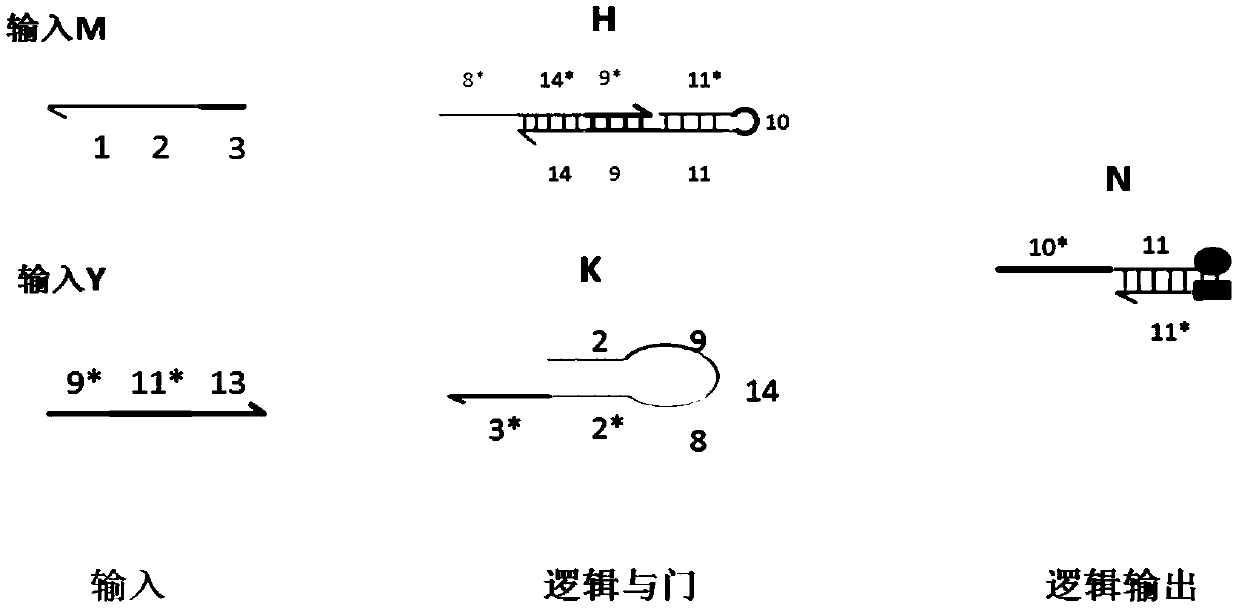
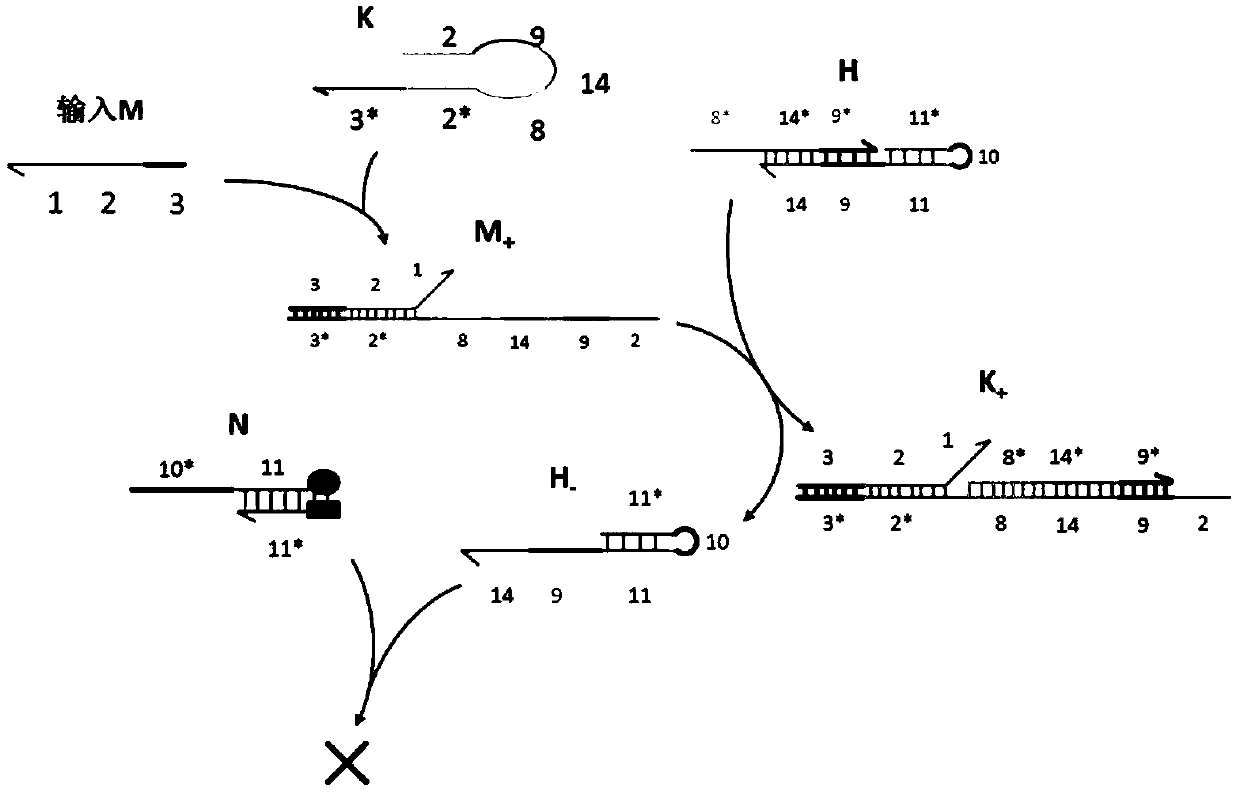
AND gate molecular circuit, NOT gate molecular circuit, XOR molecular circuit and half-subtracter molecular circuit based on DNA hairpin structure
The present invention relates to an AND gate molecular circuit, a NOT gate molecular circuit, an XOR molecular circuit and a half-subtracter molecular circuit based on a DNA hairpin structure, belonging to the biocomputer technology field. The principles of the DNA single hairpin structure, the double hairpin structure and the two-way strand displacement reaction are employed to design a strand displacement reaction structure taking a conventional small fulcrum as an identification area to realize the construction of AND gate, NOT gate and XOR gate basic logic operation structures and perform globality simulation argumentation. A basic logic circuit is employed to perform combination design of the half-subtracter molecular circuit and perform argumentation simulation of the circuit feasibility.
Owner:ZHENGZHOU UNIVERSITY OF LIGHT INDUSTRY
Application of Docker technology in high-throughput sequencing data analysis
InactiveCN107590365ABreak down complexityBreak the environment dependencySpecial data processing applicationsComputer resourcesData information
The invention discloses an application of a Docker technology in high-throughput sequencing data analysis, belonging to the field of biocomputing and molecular biology. The application comprises the following steps: firstly, constructing a base image by using a Docker, and storing software and languages required for analysis into the image in an interactive mode; building an analysis process, andthen generating a biological cloud computing platform image; saving the biological cloud computing platform image as a tar compression package, uploading and importing to a server directory that needsto be analyzed, and then mounting data information and data annotation files that need to be analyzed to the image, and calling the configured analysis process to analyze data. According to the application disclosed by the invention, the Docker technology is applied to high-throughput sequencing data analysis, the problem of limitations in software installation, configuration, migration, the resource difference of dependent computers and the like in existing biological big data analysis can be solved, and thus researchers can efficiently mine and analyze biological sequencing big data, and the time of processing analysis methods can be reduced.
Owner:武汉古奥基因科技有限公司
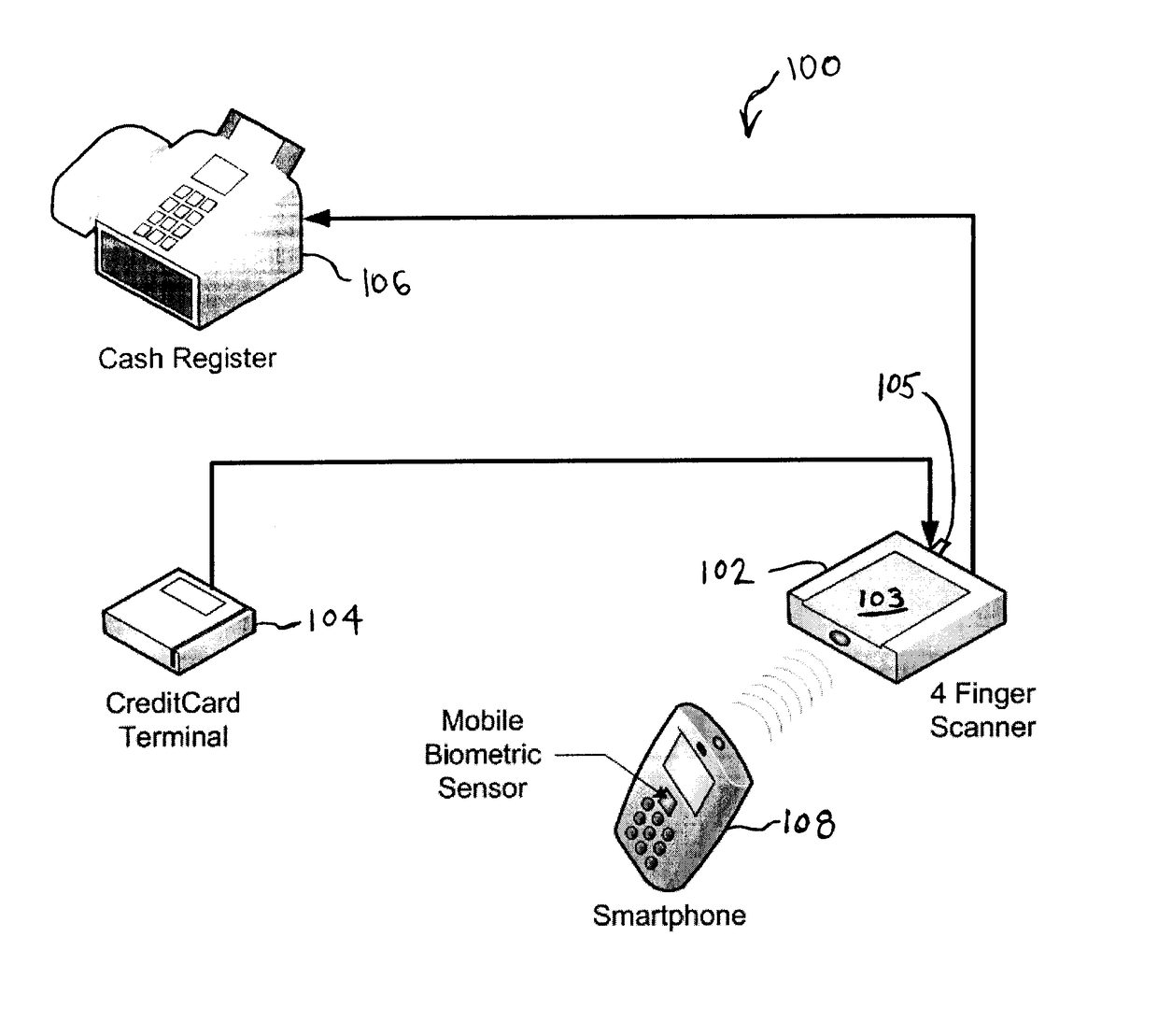
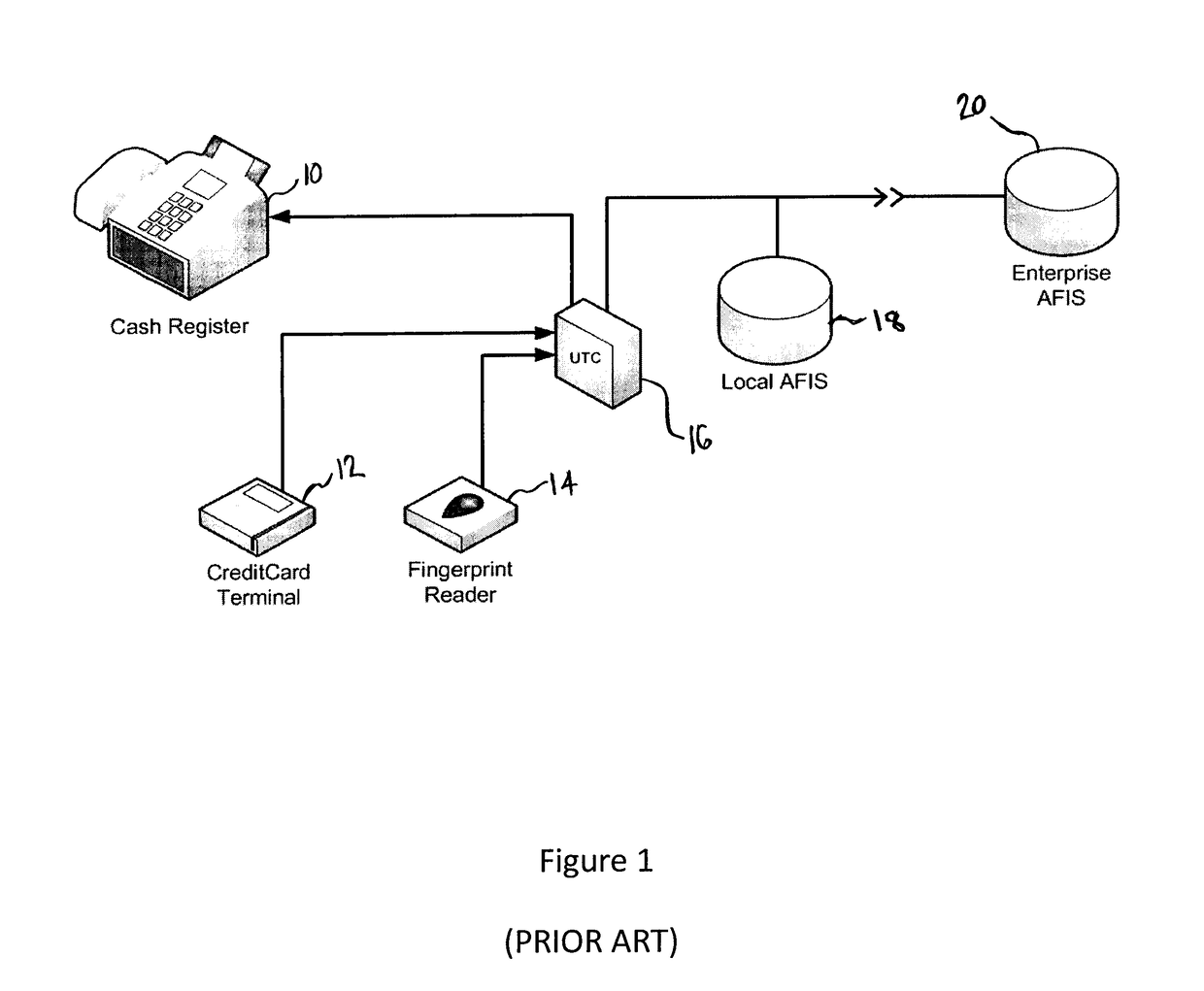
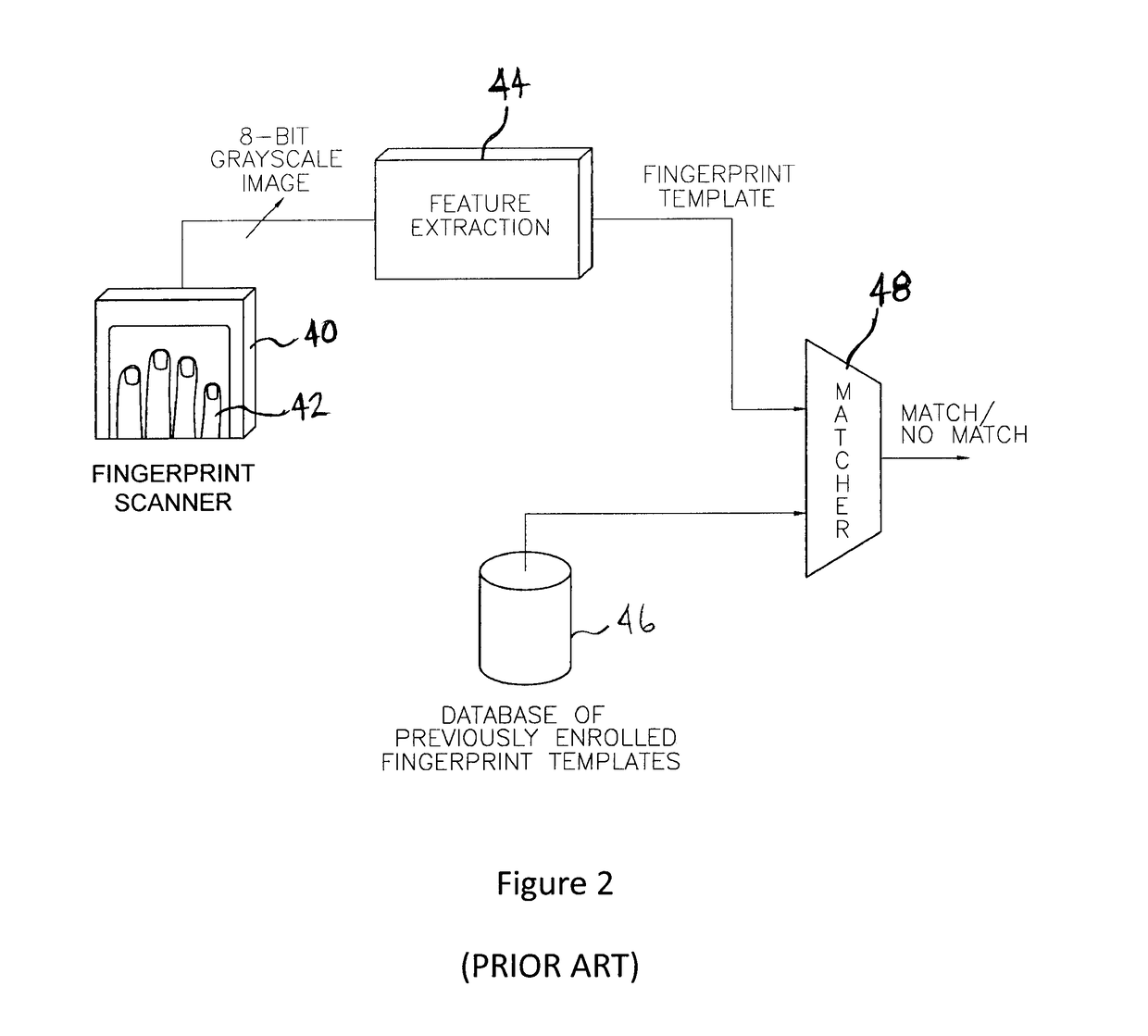
Biometric computing system and method for e-commerce
A system and method for authorizing transactions using biometrics is disclosed. The system may be a commerce authorization system comprising a portable computing device, a docking station, and a biometric reader. The method for authorizing a transaction may comprise the steps of: receiving biometric data from a biometric reader; comparing the received biometric data with a biometric template; and, transmitting payment information to the vendor.
Owner:QUALCOMM INC
Microfluidic xor operation system and method based on molecular beacon DNA
PendingCN109055493ASafe storageGuaranteed input combinationMicrobiological testing/measurementSpecial data processing applicationsDNA SolutionsEngineering
The invention discloses a microfluidic xor operation system and method based on molecular beacon DNA in the technical field of biological computers. The system comprises a microfluidic chip, wherein the microfluidic chip is provided with a first input signal chamber and a second input signal chamber, a first microfluidic channel is arranged between the first input signal chamber and the second input signal chamber, an S-shaped channel is connected with the right end of the first microfluidic channel, a second microfluidic channel is formed in the right side of the S-shaped channel, and the right end of the second microfluidic channel is connected with a liquid storage chamber. Two input signal chambers are arranged, so that DNA solutions representing two input signals can be stored respectively, and four input combinations can be ensured; the operation system is arranged on the microfluidic chip, so that flow direction control of fluid can be realized, the temperature required for reaction and a detection operation result can be provided, operation is simple, and accuracy is high; DNA calculation is combined with the microfluidic technology, the integration level is high, and operation is fast and accurate.
Owner:ANHUI UNIVERSITY +1
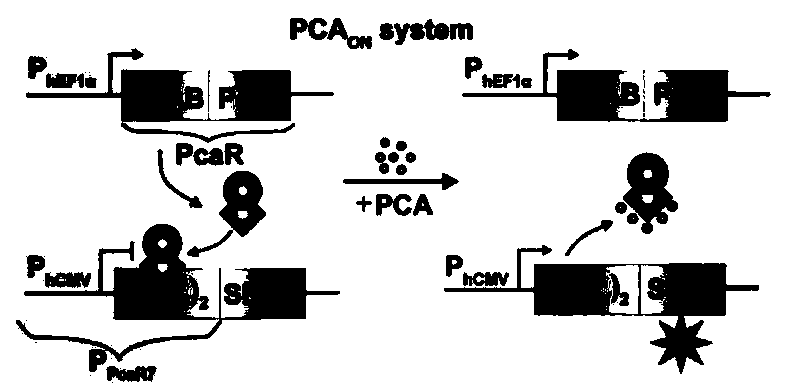
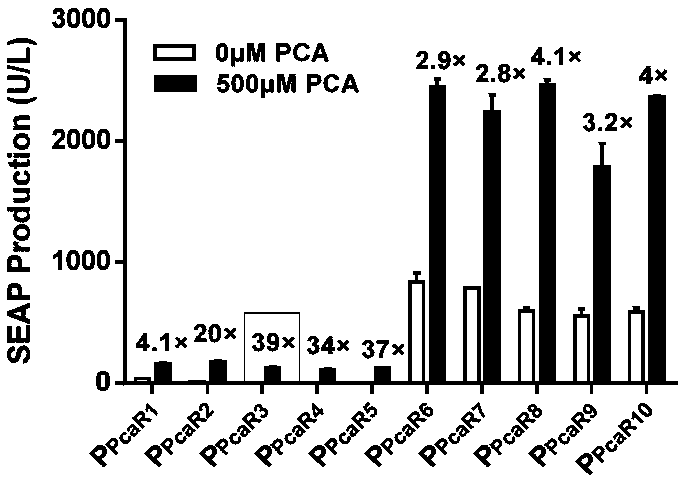
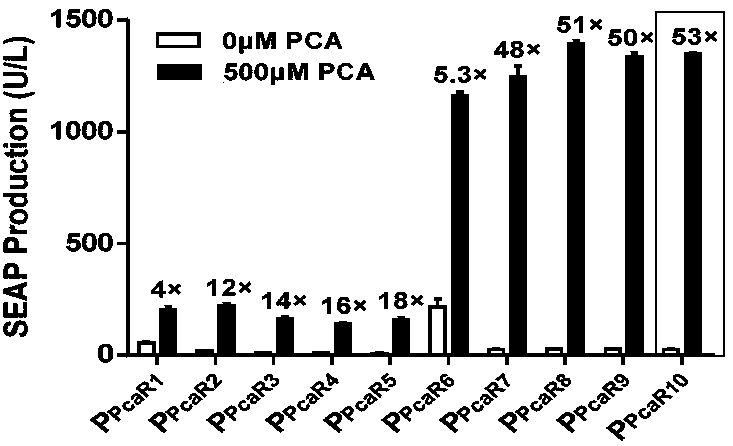
Protocatechuic acid regulated on-off system as well as regulation method and application thereof
ActiveCN109456991AGood hypoglycemic effectMetabolism disorderStable introduction of DNAMetaboliteRegulation of gene expression
The invention discloses a multifunctional gene expression platform regulated by green tea metabolite protocatechuic acid. The platform relates to an on-off system regulated by protocatechuic acid, i.e. the expression is initiated or closed by the protocatechuic acid regulation gene. The on-off system comprises a protocatechuic acid regulated 'on' system and a protocatechuic acid regulated 'off' system. The invention also provides a eukaryotic expression vector comprising the on-off system, a mammalian cell line and a microcapsule. The invention also discloses an application of the multifunctional platform capable of the accurate expression release of controllable insulin or glucagon-like peptide in treating diabetes; and the gene expression regulation platform can be used for establishinga complicated biological computer as well as regulating the gene edition and gene expression activation or inhibition mediated by a CRISPR / Cas9 or CRISPR / dCas9 system. The multifunctional gene expression platform provides a useful novel gene expression regulation tool for the gene treatment and cell treatment.
Owner:EAST CHINA NORMAL UNIV
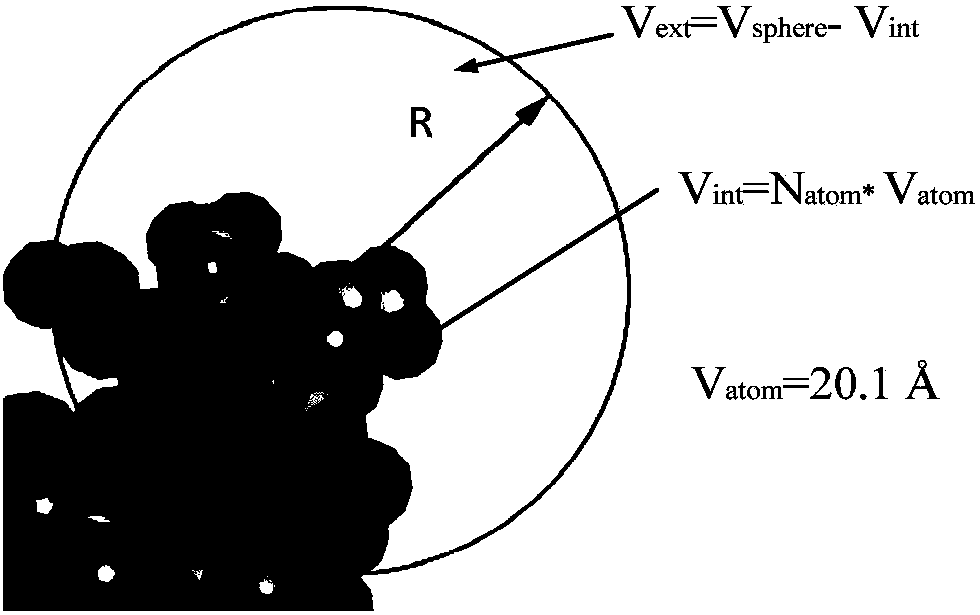
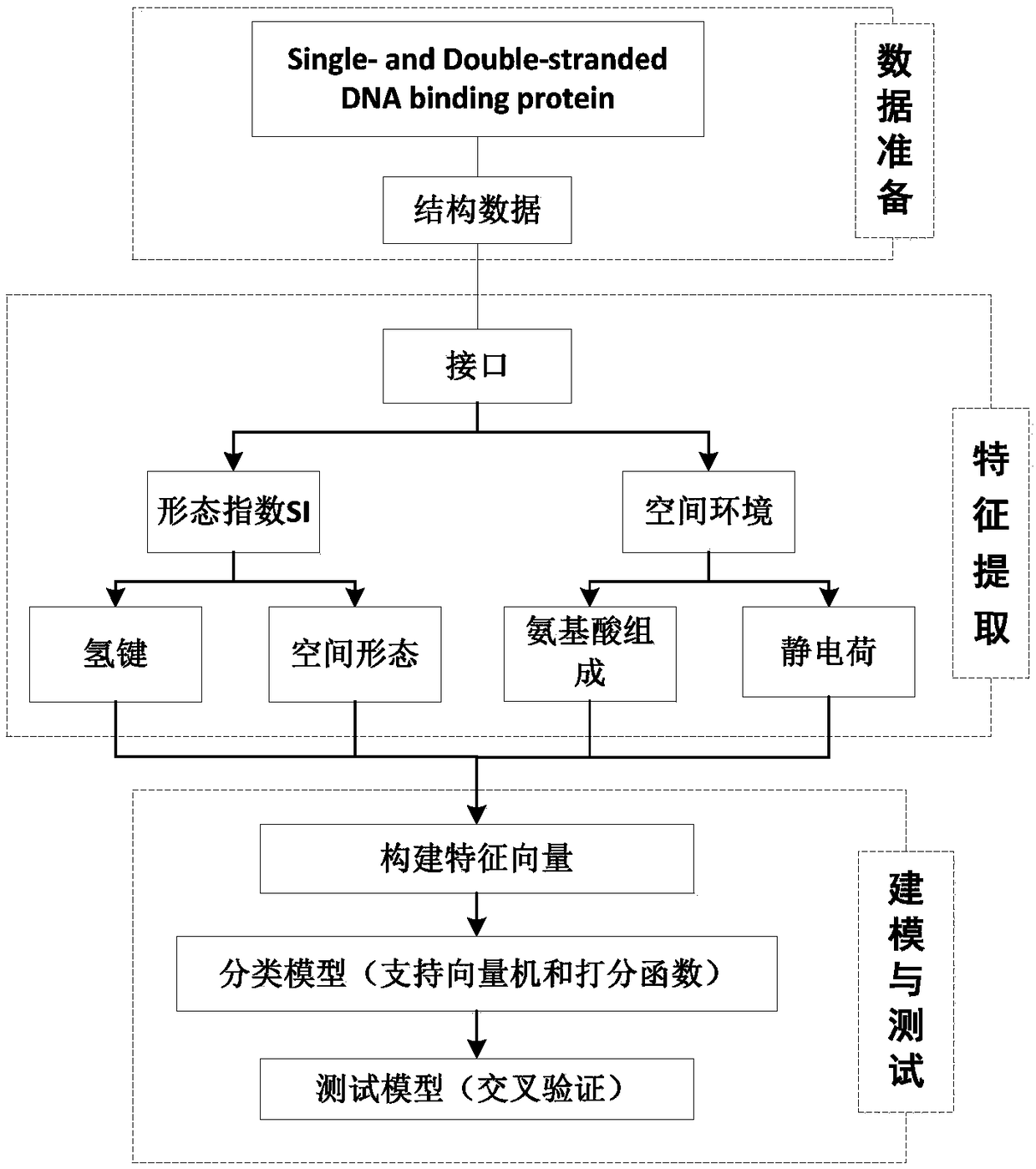
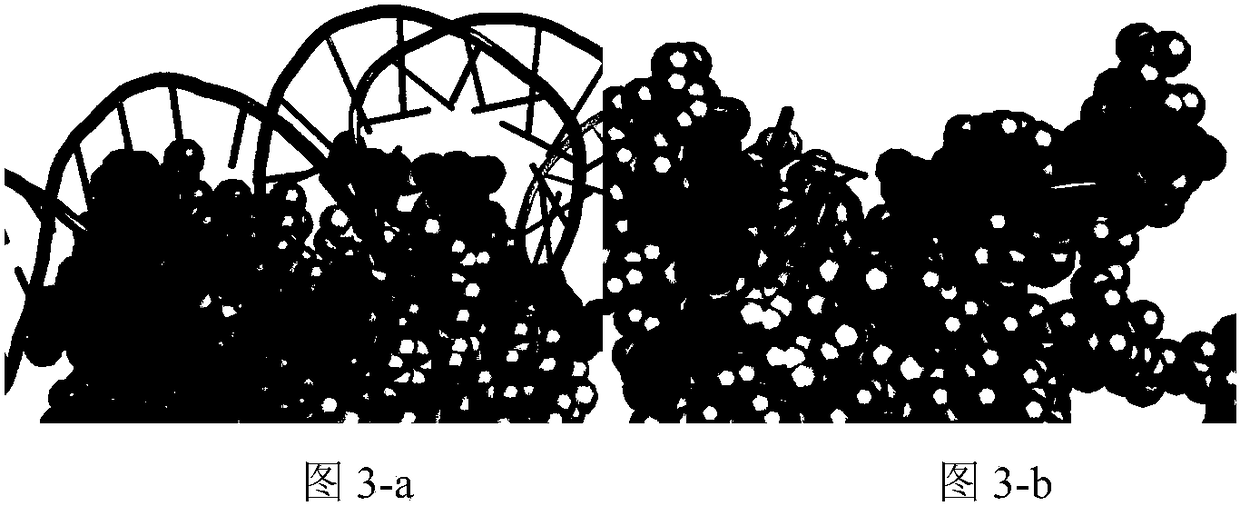
A method for analyzing the geometric structure characteristics of DNA bin protein interface
InactiveCN109101784AIn-depth understanding of the binding mechanismSpecial data processing applicationsBinding siteSingle strand dna
The invention relates to an analysis method for the geometrical structure characteristics of DNA binding protein interface, belonging to the technical field of biological computing. The present invention first derives proteins-DNA binding site information from proteins-DNA complex dataset; and then based on the protein-DNA Binding Site Information, residue Morphological Index and Spatial Environment of proteins-DNA Binding Sites are determined; finally, the morphological index and spatial environment of the identified protein-DNA binding sites are used as feature vectors to classify the obtained feature vectors by using classification model. As the binding specificity of double-stranded DNA binding protein (DSBs) and single-stranded DNA binding proteins (SSBs) is analyzed from the angles of the morphological structure of the residues and the characteristics of the space environment, the invention is conducive to the in-depth understanding of the proteins, and the binding specificity ofthe double-stranded DNA binding proteins and the single-stranded DNA binding proteins is analyzed. Based on the DNA binding mechanism, the present invention can also be applied to proteins-RNA, protein-Protein and other fields.
Owner:HENAN NORMAL UNIV
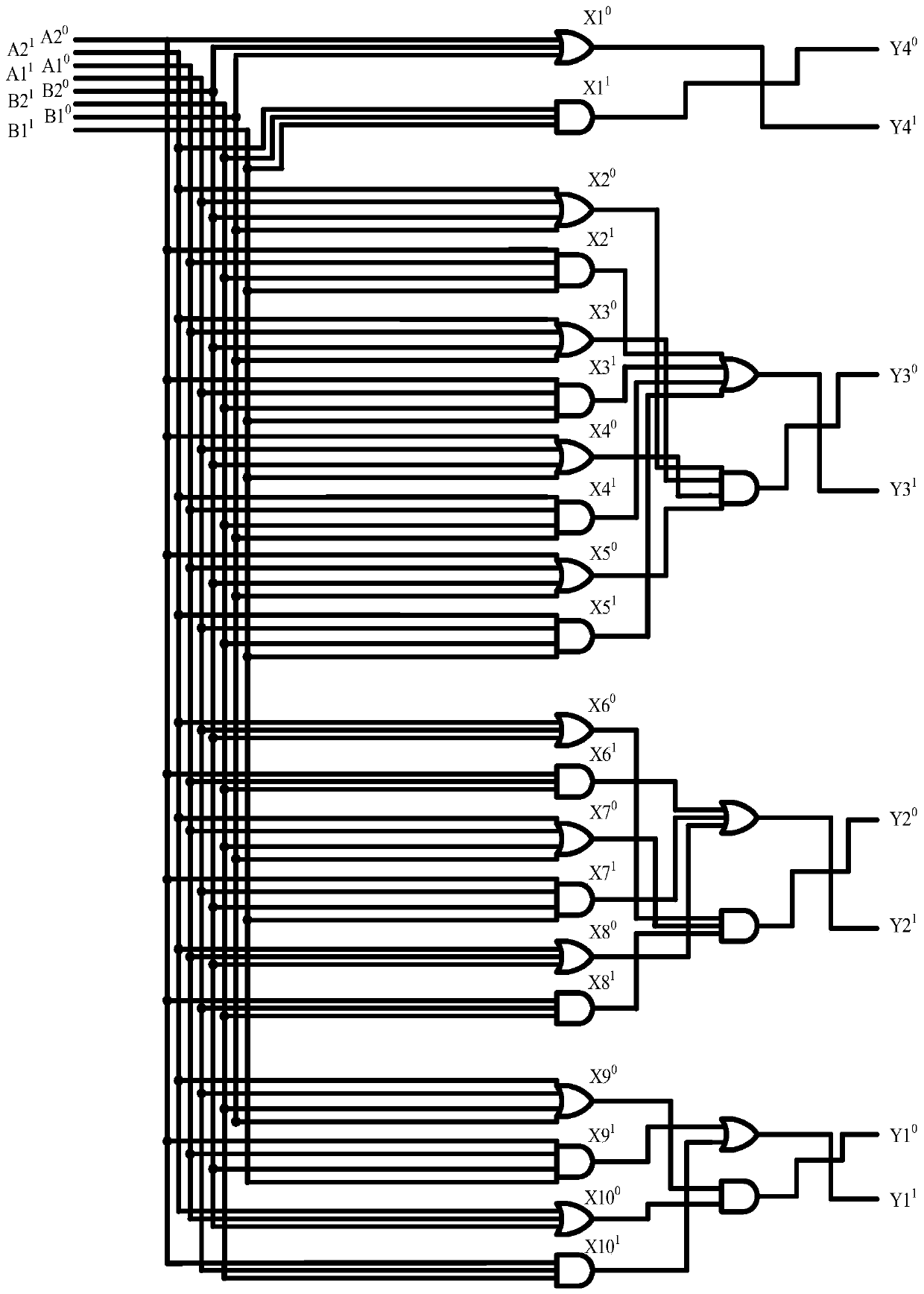
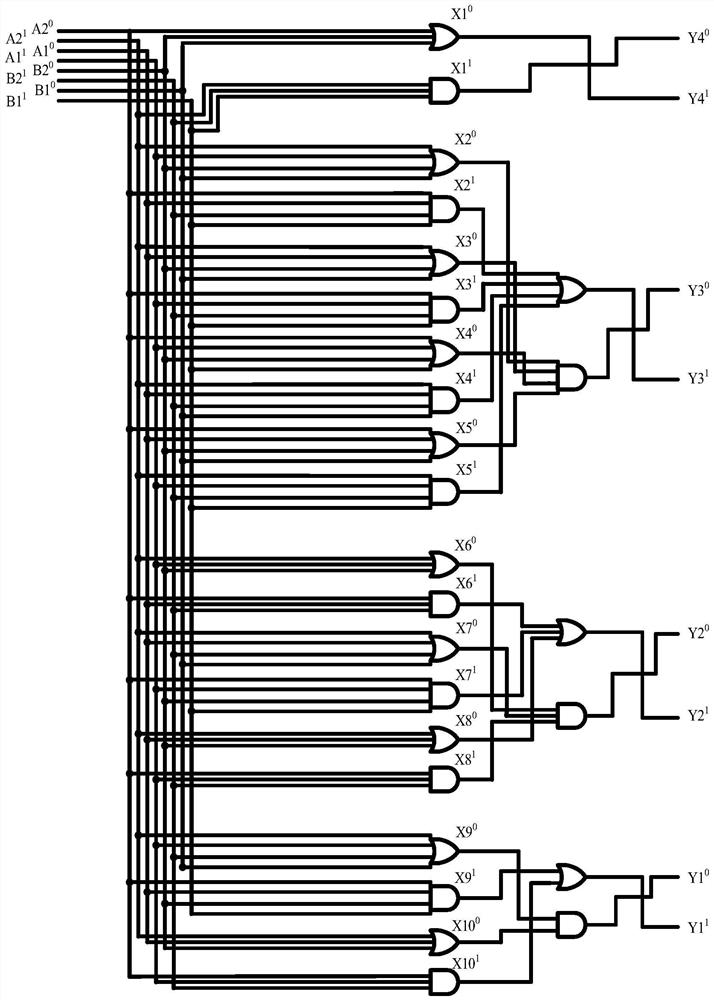
Four-input factorial additive operation molecular circuit design method based on DNA strand displacement
ActiveCN110544511AImprove reliabilityComputation using non-contact making devicesSpecial data processing applicationsComputer architectureRandom combination
The invention provides a four-input factorial addition operation molecular circuit design method based on DNA strand displacement, and the method comprises the following steps: listing factorial multiplications corresponding to two binary numbers, listing a truth table of the sum of the factorial multiplications of ten kinds of two binary numbers according to a random combination, and constructinga factorial addition operation circuit; converting a four-input factorial additive operation circuit into a double-track logic circuit only comprising a logic AND gate and a logic OR gate by adoptinga double-track idea; designing a DNA molecular logic gate through DNA molecules, constructing a four-input fractional addition operation molecular circuit, verifying the correctness of an output result through Visual DSD simulation software, analyzing complex dynamic behaviors of the four-input fractional addition operation molecular circuit, and verifying the dynamic behaviors of the four-inputfractional addition operation molecular circuit. According to the invention, a basic theoretical basis is provided for constructing a more complex logic operation circuit in the future, and the development of the biological computer is promoted, so that the reliability of the logic circuit of the biological computer is improved.
Owner:ZHENGZHOU UNIVERSITY OF LIGHT INDUSTRY
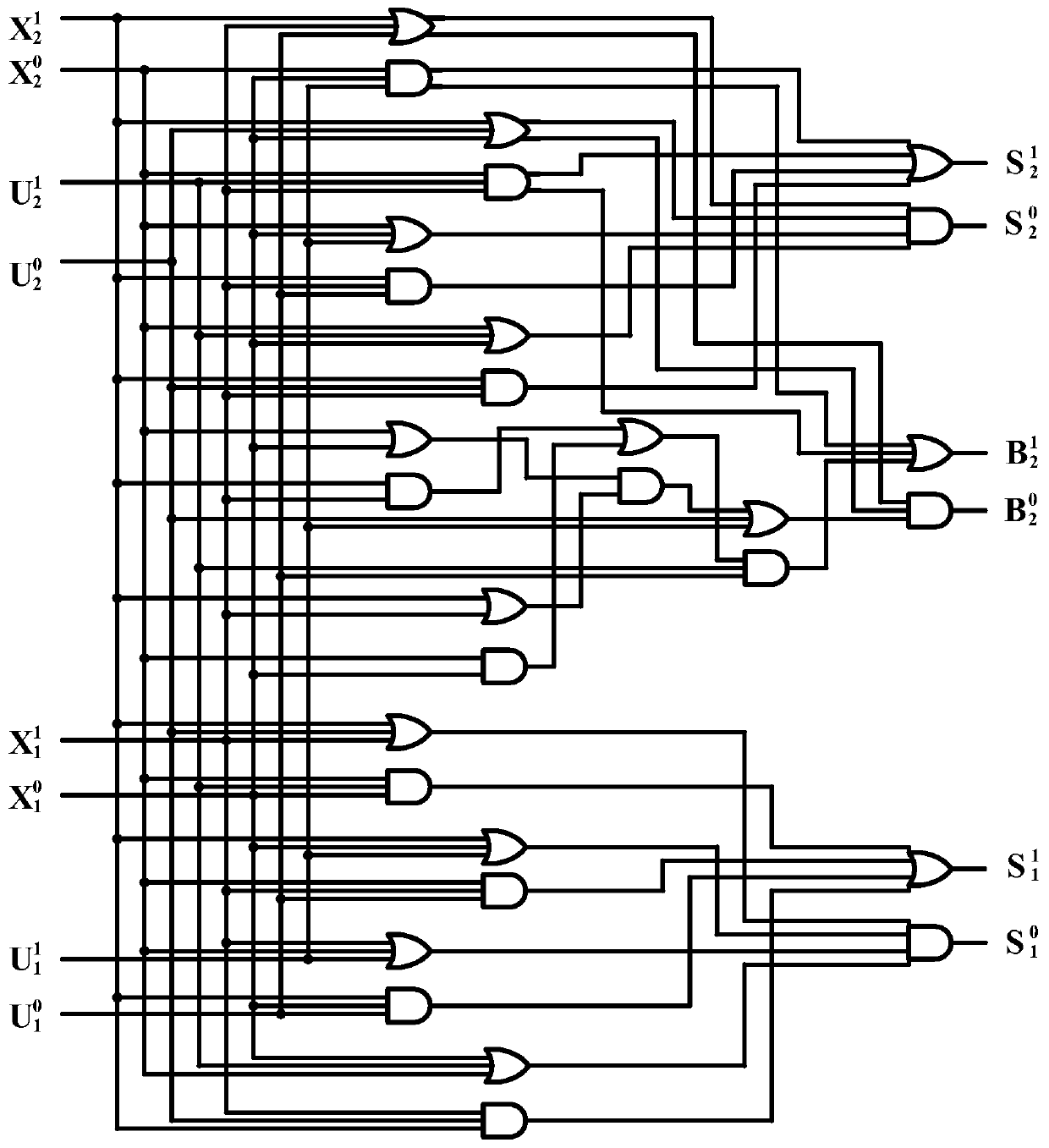
Implementation method of two-bit Gray code subtractor molecular circuit based on DNA strand displacement
ActiveCN110532705AImprove propulsion performanceImprove reliabilitySpecial data processing applicationsDisplacement reactionsTruth table
The invention provides an implementation method of a two-bit Gray code subtractor molecular circuit based on DNA strand displacement. The implementation method is used for solving the problem of two-bit Gray code subtractor molecular operation based on Gray codes. The method comprises the following steps of: listing an operation truth table according to an operation rule of a two-bit Gray code subtractor circuit; converting the two-bit Gray code subtractor circuit into a two-bit Gray code subtractor double-track logic circuit by using a double-track idea; researching a DNA strand displacementreaction principle based on DNA strand displacement reaction; designing and cascading all DNA molecule fan-out doors, DNA molecule AND gates, DNA molecule OR gates and DNA report gate structures according to a DNA strand displacement reaction principle, and constructing a two-bit Gray code subtractor molecular circuit; visual DSD software is used for carrying out simulation analysis on the molecular circuit, and the dynamic characteristics of the molecular circuit are verified. According to the invention, a basic theoretical basis is provided for constructing a more complex logic operation circuit in the future, and the development of the biological computer is promoted, so that the reliability of the logic circuit of the biological computer is improved.
Owner:ZHENGZHOU UNIVERSITY OF LIGHT INDUSTRY
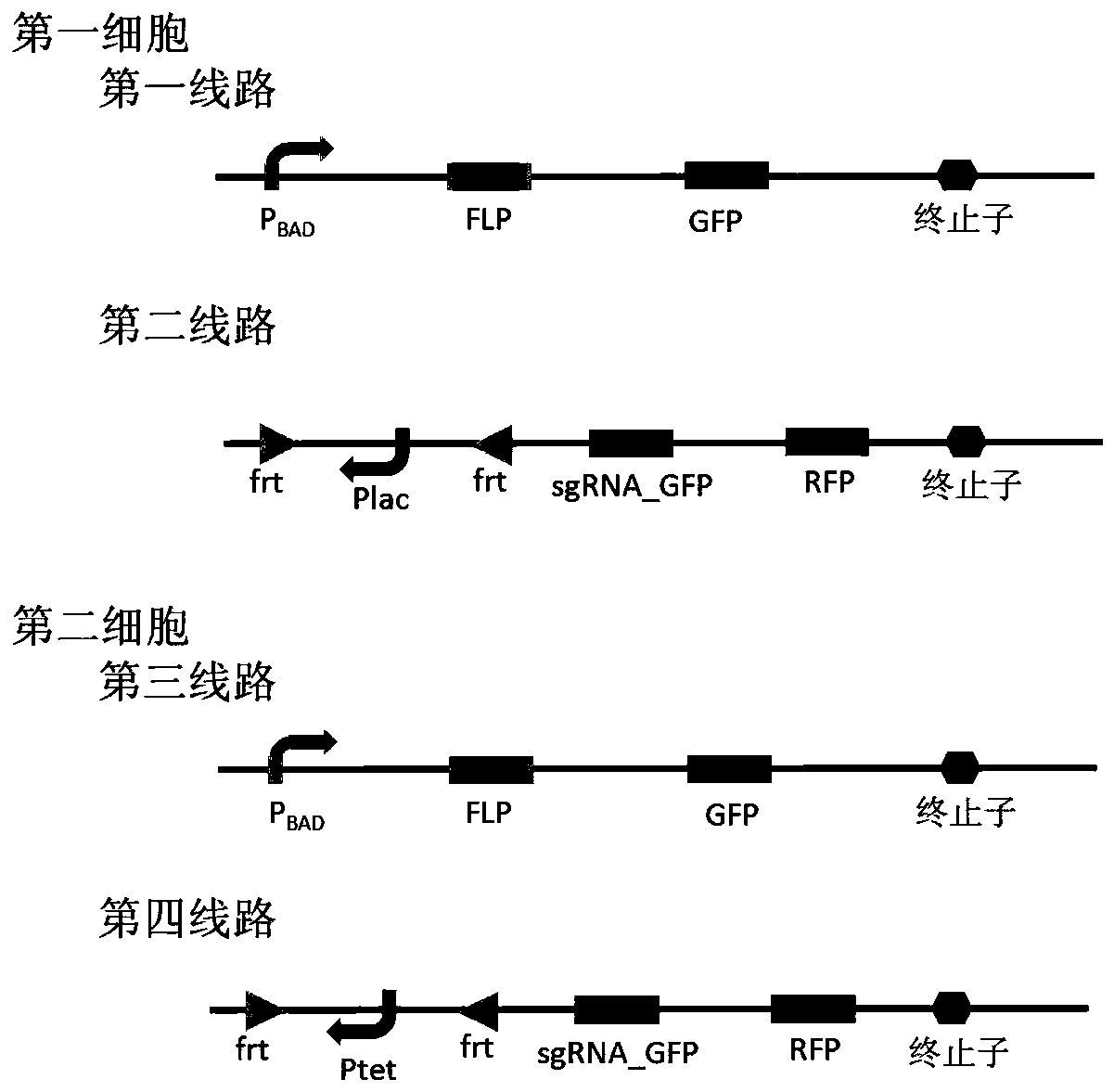
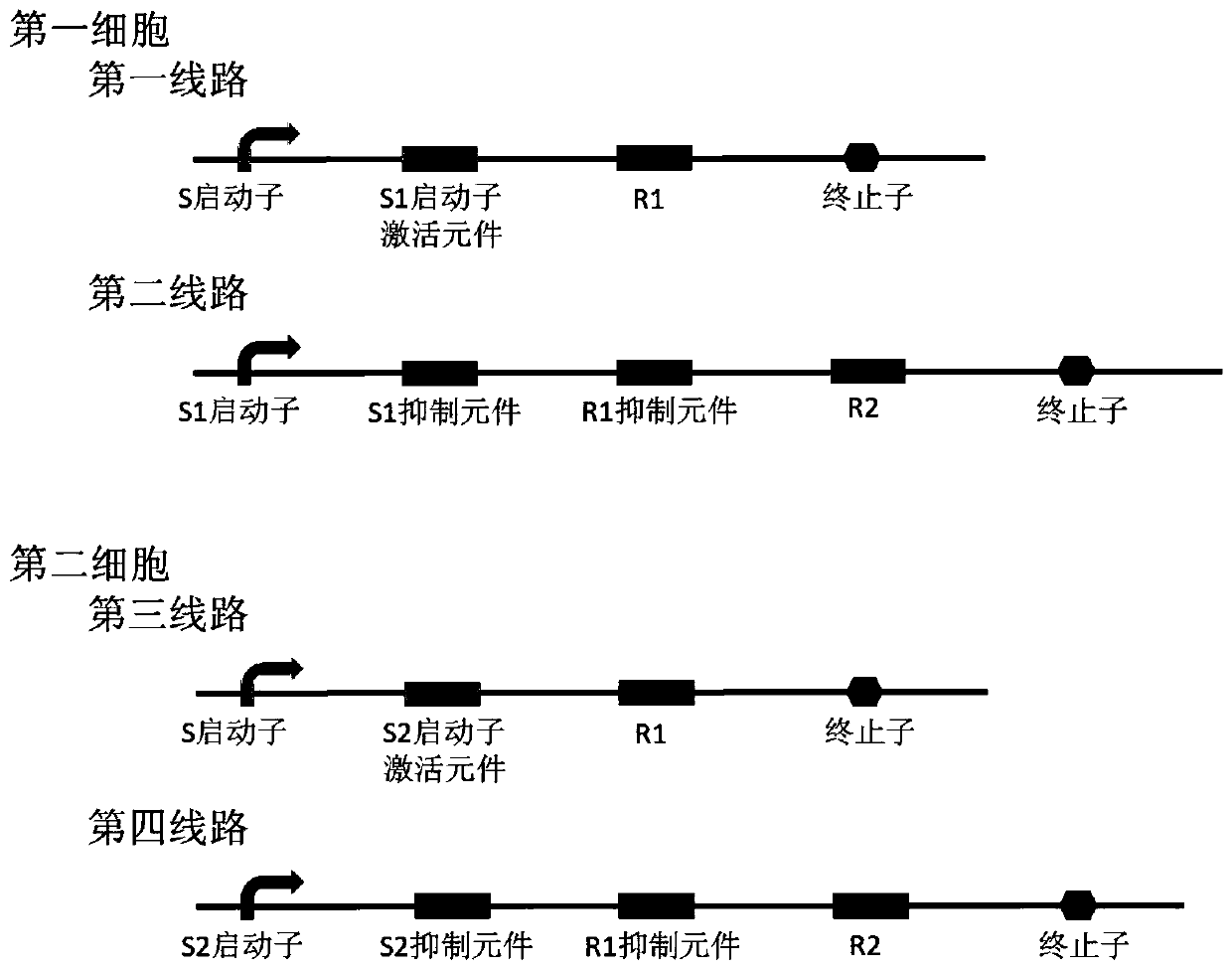
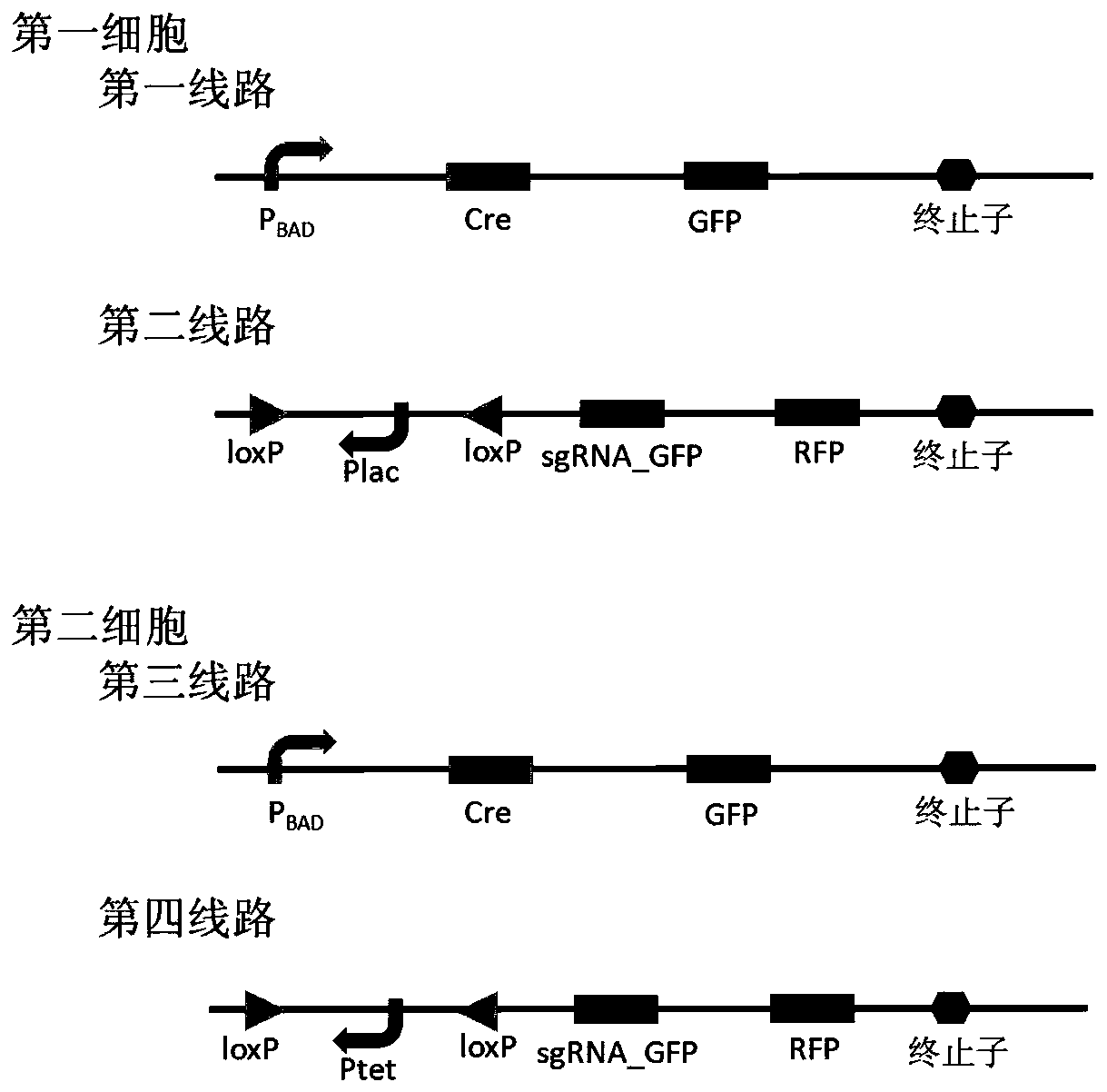
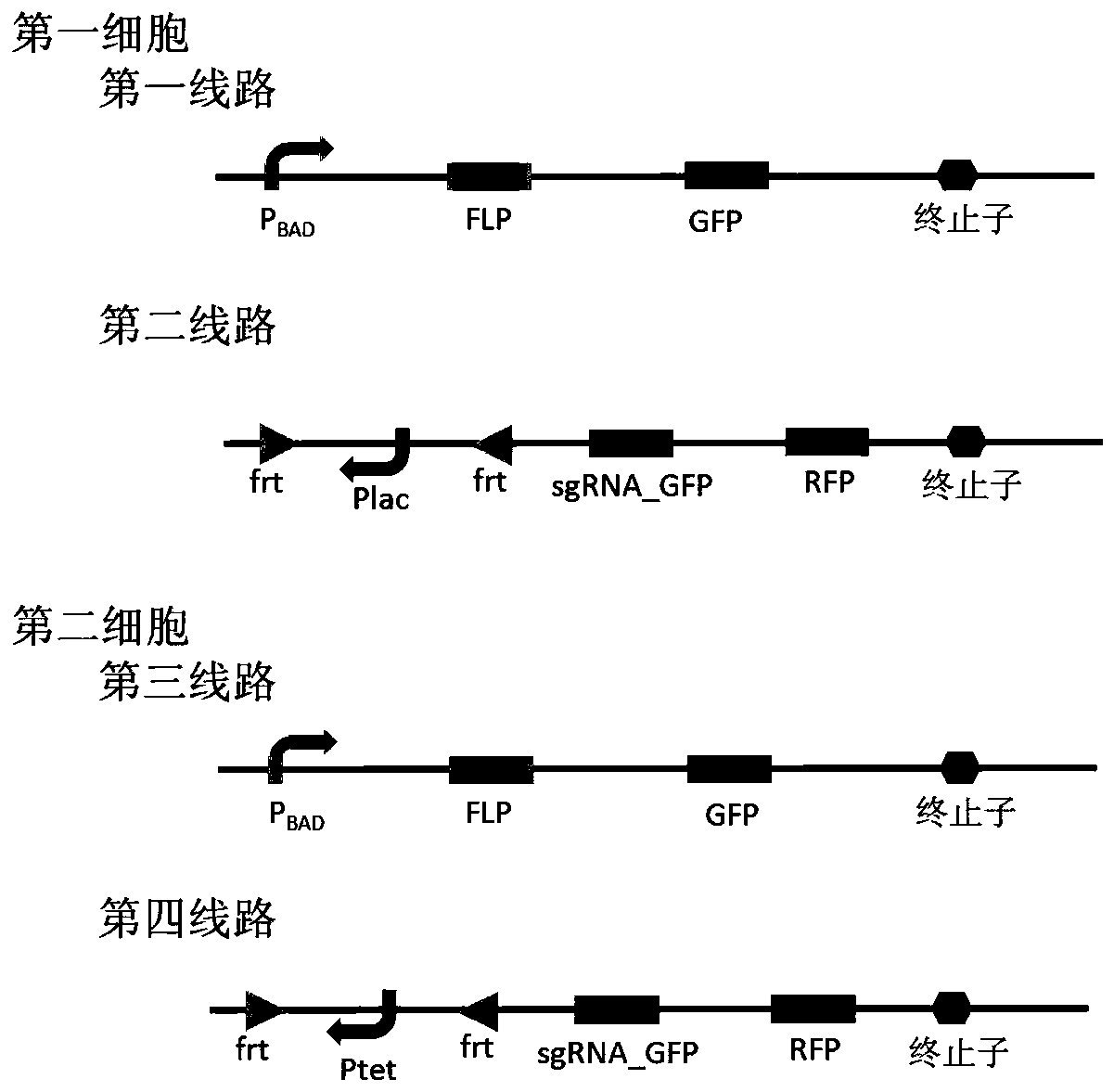
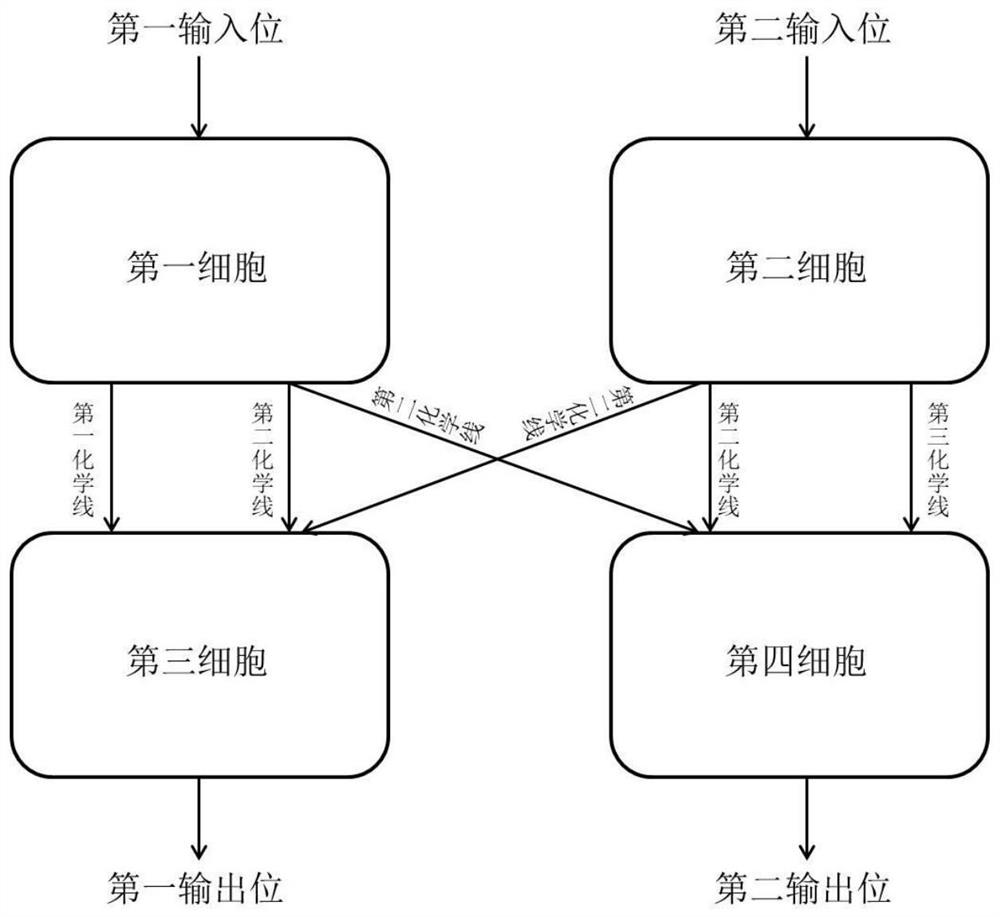
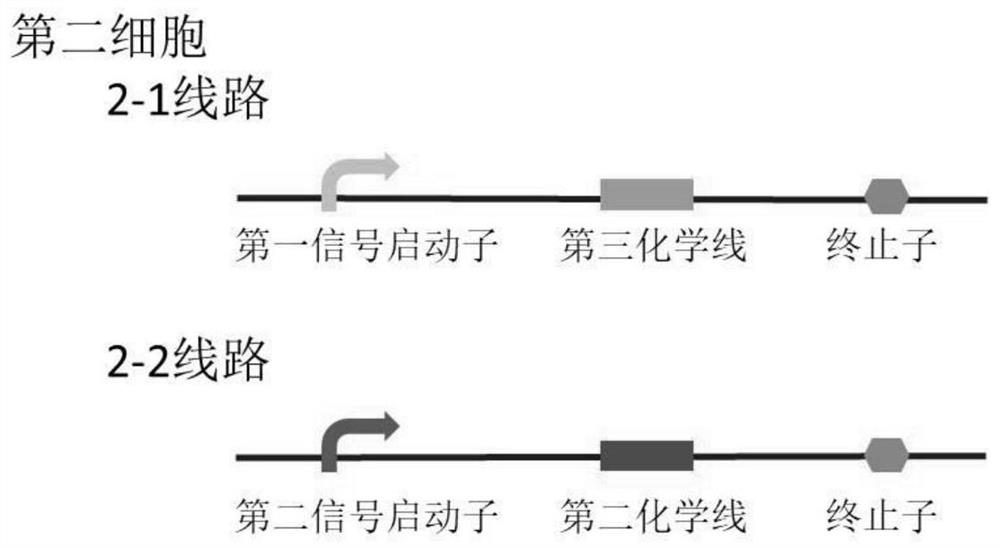
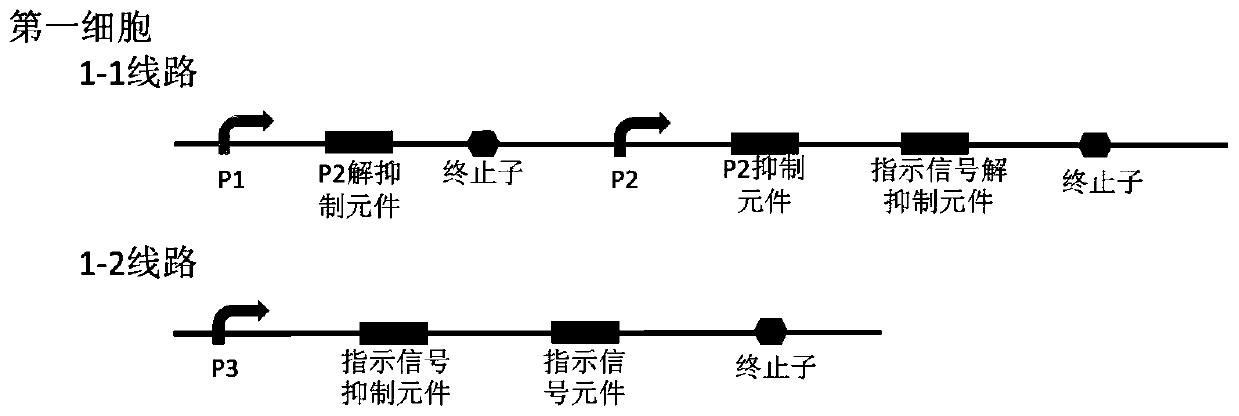
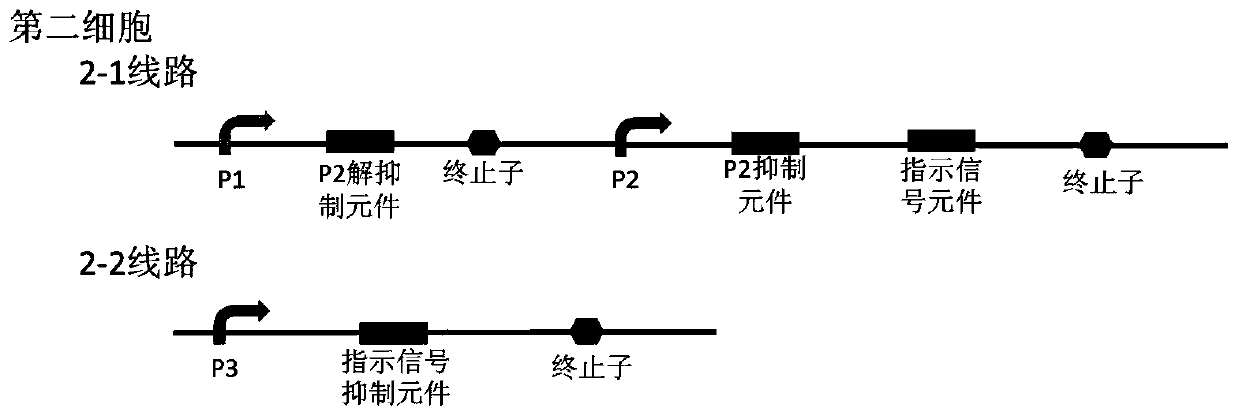
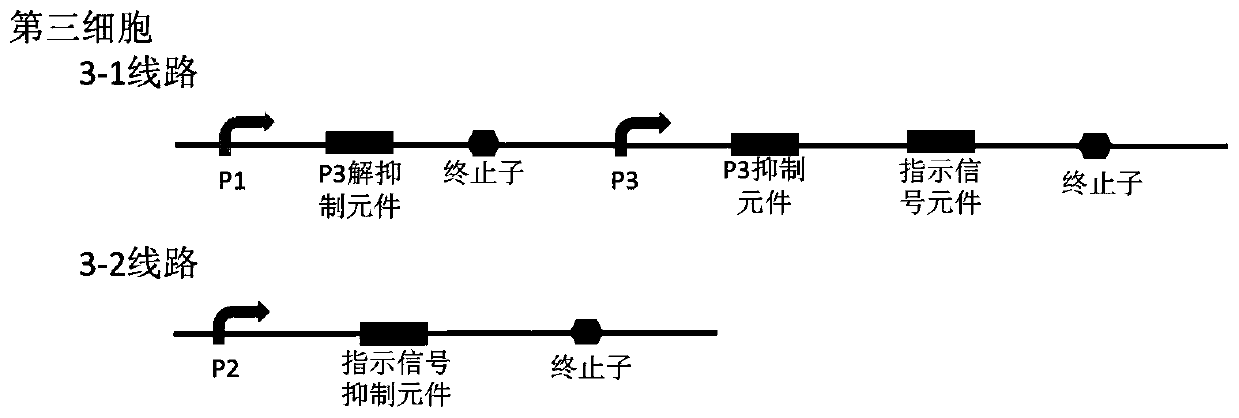
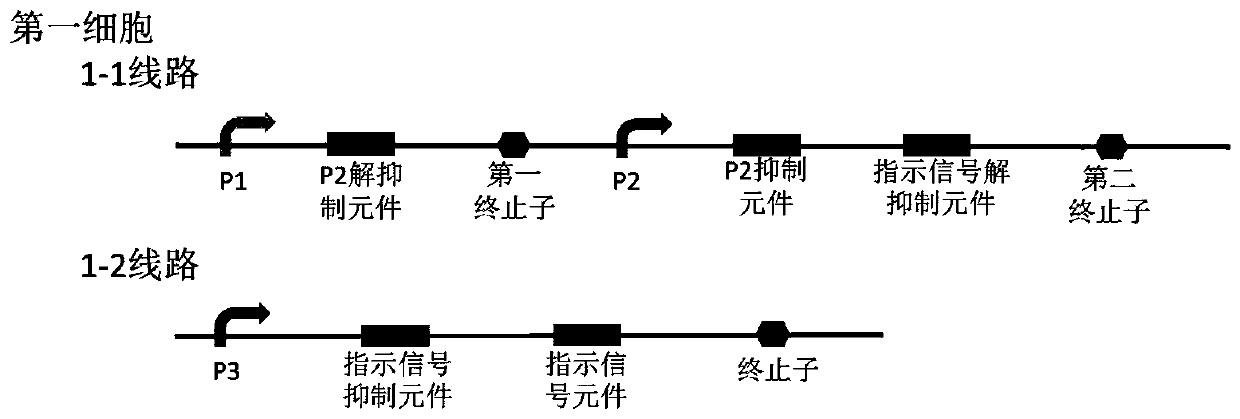
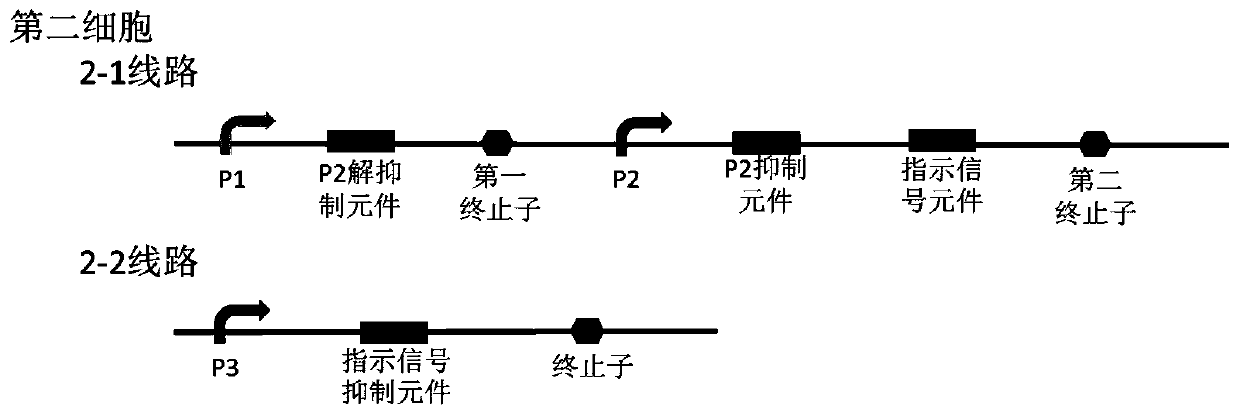
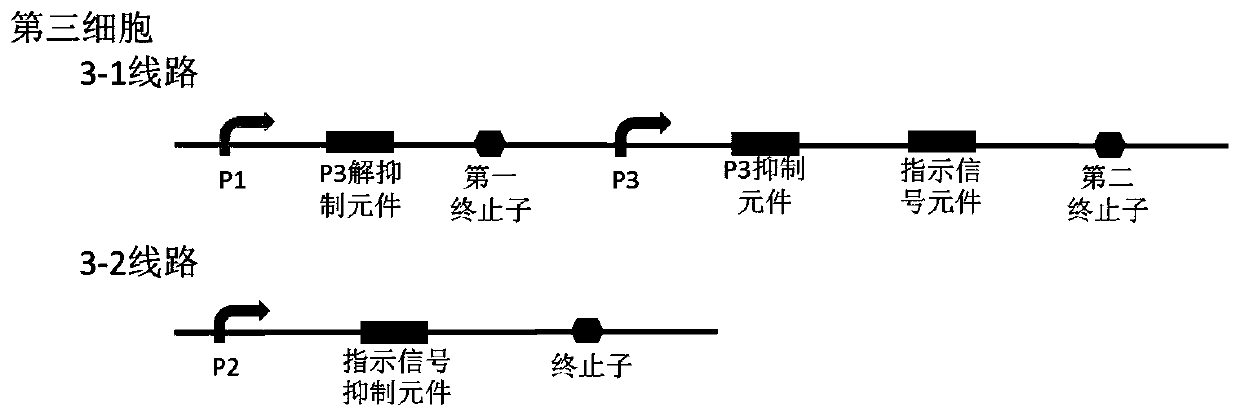
Bacterial cell computing component that indicates priority of a signal at different accompanying signals
The present disclosure provides a bacterial cell computing component that indicates priority of a signal at different accompanying signals. A cell calculation part is composed of a first cell and a second cell. The first cell is used for detecting a signal to be detected and a first accompanying signal, and the second cell is used for detecting the signal to be detected and a second accompanying signal; the gene lines in the first cell comprise a first line and a second line, and the gene lines in the second cell comprise a third line and a fourth line. According to the cell calculation part provided by the invention, by designing a gene line, activation of the promoter and inhibition of the indication signal are used for making different indications on no signal to be detected, only the signal to be detected, the signal to be detected and the corresponding accompanying signal exist at the same time, and a final result is indicated through output combination of multiple cells. The component can be used for related applications such as a biological computer and a biosensor which need to judge the priority of a certain specific signal in different states, and has the advantages of calculation accuracy, high efficiency and usability.
Owner:MINZU UNIVERSITY OF CHINA
SiRNA and expression carrier for inhibiting human telomerase reversed transcriptive enzyme gene expression and their pharmaceutical use
InactiveCN1580260AReduce degradationGrowth inhibitionOrganic active ingredientsGenetic material ingredientsTelomeraseEnzyme Gene
The present invention discloses SiRNA and expression carrier thereof for specifically inhibiting human telomerase enzyme reversed transcriptive enzyme gene expression and their pharmaceutical use for preparing human telomere enzyme activation related neoplastic medicines. The present invention obtains a group of human telomere enzyme inverse transcriptase gene sequences from the GenBank by using bio-computer information technology, based on the RNA disturbance technology, works out a group of siRNA which probably induce RNA disturbance, and expresses a certain amount of siRNA though chemical method synthesis or plasmid carrier, and then specifically restrains the expression of human telomere enzyme inverse transcriptase genes. Said siRNA and expression plasmid thereof can prepare highly effective and rapid antineoplastic drugs with strong specificity and low side-effects.
Owner:徐根兴
Short interference ribonucleic acid as novel anti-tumor gene therapeutic medicine
InactiveCN1225286COrganic active ingredientsGenetic material ingredientsHuman tumorComputers technology
By means of biological computer technology, one group of gene sequence relevant to human tumor is obtained from GenBank. Based on RNA interference technology, one group of siRNA capable of inducing RNA interference is designed. Certain amount of siRNA is synthesized chemically and used as novel efficient and specific antitumor medicine. MTT detection shows that siRNA has obvious cytotoxic effect on MCF-7, BEL 7402, KB, HT-29 and other tuomr cells. Western Blotting detection shows that siRNA can lower the expression level of specific protein by 80-90%. Morphological observation shows that siRNA can cause tumor cell produce corresponding morphological change and chromatin concentration. Compared with available technology, siRNA is one antitumor medicine with completely new mechanism, high efficiency and fast speed.
Owner:MEDICINE & BIOENG INST OF CHINESE ACAD OF MEDICAL SCI
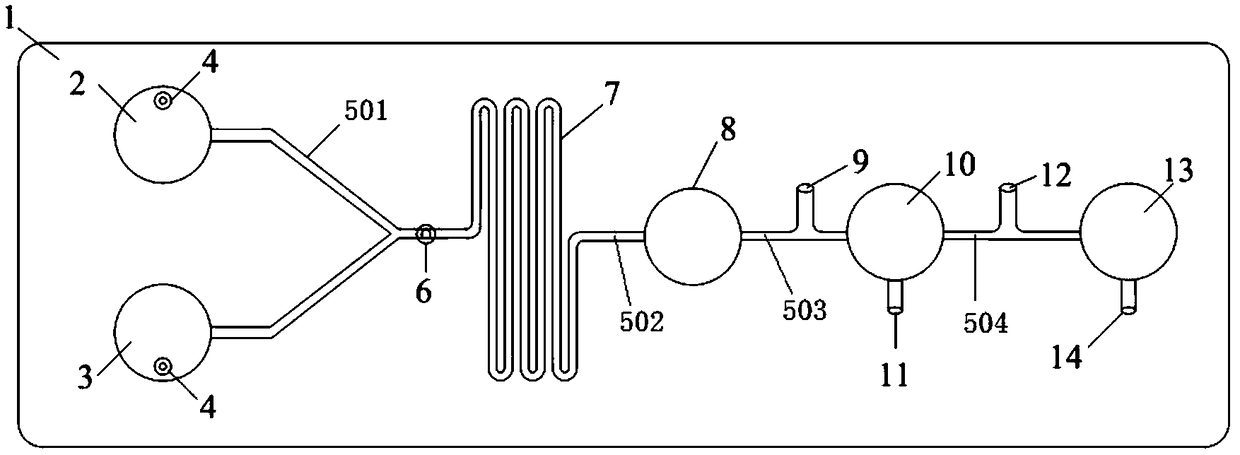
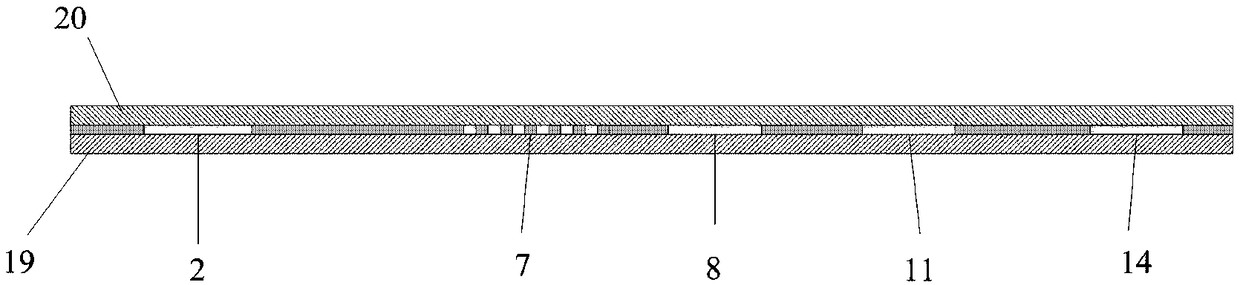
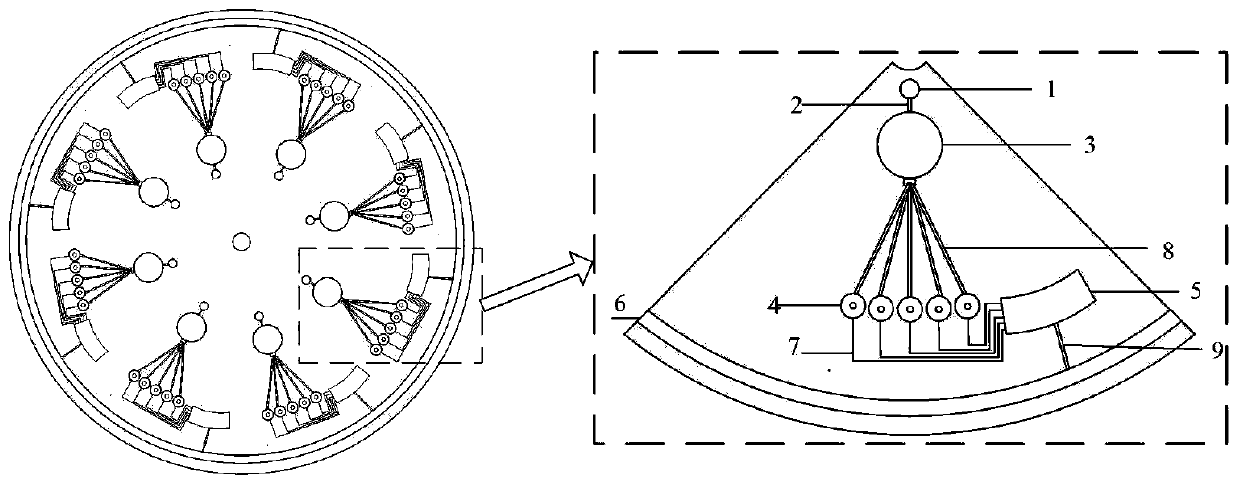
Micro-fluidic chip for solving minimum set coverage problem and DNA calculation method of micro-fluidic chip
ActiveCN111420718ASimple structureEasy to manufactureLaboratory glasswaresFluid controllersProcess engineeringA-DNA
The invention discloses a micro-fluidic chip for solving a minimum set coverage problem and a DNA calculation method of the micro-fluidic chip, and belongs to the technical field of biological computers. The micro-fluidic chip comprises a sample adding hole, a sample adding channel, a substrate liquid pool, a reaction pool group, a waste liquid pool, a liquid outlet channel, a capillary channel, afirst micro-fluidic channel and a second micro-fluidic channel, is simple in structure and convenient to manufacture, and does not need to integrate any micro-valve micro-pump and other elements; according to the method, centrifugal force is used for driving a solution on the chip to flow; all possible results can be exhausted on the chip; verifying whether a result meets a condition or not at the same moment is realized; the DNA sequence does not need to be repeatedly added on the chip for many times; compared with the prior art, manual intervention is reduced, the problems that in a traditional algorithm, the serial calculation amount is large, and time complexity is large are solved, the strong parallel capacity of DNA calculation is achieved, the MSC problem is solved through the reaction that RNA is split through DNAzyme on a chip, and the calculation reliability is effectively improved.
Owner:ANHUI UNIVERSITY +1
Biological computing systems and methods for multivariate surface analysis and object detection
PendingUS20210319279A1Improve abilitiesHigh viscosityBiomolecular computersDNA computersDiseaseEngineering
The present invention provides biological computing systems comprising computing units that process input signals to produce an output signal. In particular, the computing units include, for example, cells and proteins that function to convert biological signals into a discernable output that provides information about a biological sample. Further provided are methods of using such biological computing systems, such as for the diagnosis of various diseases and conditions.
Owner:XENO CELL INNOVATIONS SRO
Bacterial cell computing component indicating the priority of signals under different accompanying signals
The present disclosure provides a bacterial cell computing component that indicates priority of a signal at different accompanying signals. A cell calculation part is composed of a first cell and a second cell. The first cell is used for detecting a signal to be detected and a first accompanying signal, and the second cell is used for detecting the signal to be detected and a second accompanying signal; the gene lines in the first cell comprise a first line and a second line, and the gene lines in the second cell comprise a third line and a fourth line. According to the cell calculation part provided by the invention, by designing a gene line, activation of the promoter and inhibition of the indication signal are used for making different indications on no signal to be detected, only the signal to be detected, the signal to be detected and the corresponding accompanying signal exist at the same time, and a final result is indicated through output combination of multiple cells. The component can be used for related applications such as a biological computer and a biosensor which need to judge the priority of a certain specific signal in different states, and has the advantages of calculation accuracy, high efficiency and usability.
Owner:MINZU UNIVERSITY OF CHINA
The Realization Method of the Molecular Circuit of Two-bit Gray Code Subtractor Based on DNA Strand Replacement
The invention provides an implementation method of a two-bit Gray code subtractor molecular circuit based on DNA strand displacement. The implementation method is used for solving the problem of two-bit Gray code subtractor molecular operation based on Gray codes. The method comprises the following steps of: listing an operation truth table according to an operation rule of a two-bit Gray code subtractor circuit; converting the two-bit Gray code subtractor circuit into a two-bit Gray code subtractor double-track logic circuit by using a double-track idea; researching a DNA strand displacementreaction principle based on DNA strand displacement reaction; designing and cascading all DNA molecule fan-out doors, DNA molecule AND gates, DNA molecule OR gates and DNA report gate structures according to a DNA strand displacement reaction principle, and constructing a two-bit Gray code subtractor molecular circuit; visual DSD software is used for carrying out simulation analysis on the molecular circuit, and the dynamic characteristics of the molecular circuit are verified. According to the invention, a basic theoretical basis is provided for constructing a more complex logic operation circuit in the future, and the development of the biological computer is promoted, so that the reliability of the logic circuit of the biological computer is improved.
Owner:ZHENGZHOU UNIVERSITY OF LIGHT INDUSTRY
A DNA Network Construction Method Based on Strand Displacement Regulation of E6 Ribozyme Function
The invention relates to a DNA network construction method for regulating the function of E6 ribozyme based on strand displacement. Based on the hairpin-like secondary structure of the E6 ribozyme and the sequence of the catalytic core conservative domain, the method realizes the regulation of the function of the E6 ribozyme through strand displacement technology; the substrate binding arm of the E6 ribozyme is used to add an RNA-modified DNA substrate, Form a branch ring structure to construct a DNA logic computing unit; construct a DNA logic gate by transforming the DNA logic computing unit; finally connect the DNA logic gate to form a DNA network. The method does not destroy the sequence integrity of the E6 ribozyme, and the DNA logic computing unit has the characteristics of stable structure, customization, modularization, low leakage, and anti-interference, and can be widely used in biological computing and DNA nanostructures.
Owner:DALIAN UNIV
Molecular circuit design method based on four-input factorial addition operation based on DNA strand replacement
ActiveCN110544511BImprove reliabilityComputation using non-contact making devicesCAD circuit designSoftware engineeringDna strand displacement
Owner:ZHENGZHOU UNIVERSITY OF LIGHT INDUSTRY
A kind of multi-cell signal size comparator, comparison method and application
The invention discloses a multi-cell signal size comparator, a comparison method and an application. The comparator includes four kinds of bacterial cells, the first cell and the second cell are input receiving cells, and the third cell and the fourth cell are result output cells; The first cell corresponds to the first input bit, the second cell corresponds to the second input bit, the third cell corresponds to the first output bit, and the fourth cell corresponds to the second output bit; between the first cell and the third cell The first chemical line or the second chemical line communicates, the second cell and the third cell communicate through the second chemical line, the first cell and the fourth cell communicate through the second chemical line, the second cell and the fourth cell They communicate with each other through the second chemical line or the third chemical line. Through multi-cell cooperation, the size judgment of the input signals of two input bits can be judged, and whether the two input bit signals are equal can be used in related applications such as biological computers that require size comparison.
Owner:MINZU UNIVERSITY OF CHINA
SiRNA and expression plasmid for inhibiting human bc1-2 gene expression and their use for preparing medicine
InactiveCN1233831CInhibitionGrowth inhibitionSugar derivativesGenetic material ingredientsChemical compositionSide effect
The invention publishes a kind of especial siRNA and expression plasmid, which can control men's siRNA gene expression, and their application in curing or preventing knub. The invention can get a group of bcl-2 gene orders from geneBank by biotechnology and bases on the RNA interference technology. By this condition, a group of siRNA which can induce RNA interference and can be gotten. And quantitative siRNA by chemical composition can be composed to control bcl-2 gene expression and to control neoplasitic growth. The siRNA and the expression plasmid can be used to produce antineoplasitic that has high active, high specificity and little side effect.
Owner:SUZHOU SIXTH PHARMA PLANT OF JIANGSU WUZHONG PHARMA GROUP
SiRAN and expression carrier for inhibiting human VEGF gene expression and their pharmaceutical use
InactiveCN100352922CGrowth inhibitionSmall action doseSugar derivativesGenetic material ingredientsOncologyBlood vessel
The invention publish a kind of siRNA and express carrier, which can control men's VEGF gene expression, and their application in preparing drug for affection relation to VEGF gene. The invention get a group VEGF orders by biocomputer technology and work out a group of siRNA to induce RNA disturbance basing on the RNA disturbance technology. After this, compose.
Owner:徐根兴
Molecular circuits of AND gate, NOT gate, XOR and half-subtractor based on DNA hairpin structure
The present invention relates to an AND gate molecular circuit, a NOT gate molecular circuit, an XOR molecular circuit and a half-subtracter molecular circuit based on a DNA hairpin structure, belonging to the biocomputer technology field. The principles of the DNA single hairpin structure, the double hairpin structure and the two-way strand displacement reaction are employed to design a strand displacement reaction structure taking a conventional small fulcrum as an identification area to realize the construction of AND gate, NOT gate and XOR gate basic logic operation structures and perform globality simulation argumentation. A basic logic circuit is employed to perform combination design of the half-subtracter molecular circuit and perform argumentation simulation of the circuit feasibility.
Owner:ZHENGZHOU UNIVERSITY OF LIGHT INDUSTRY
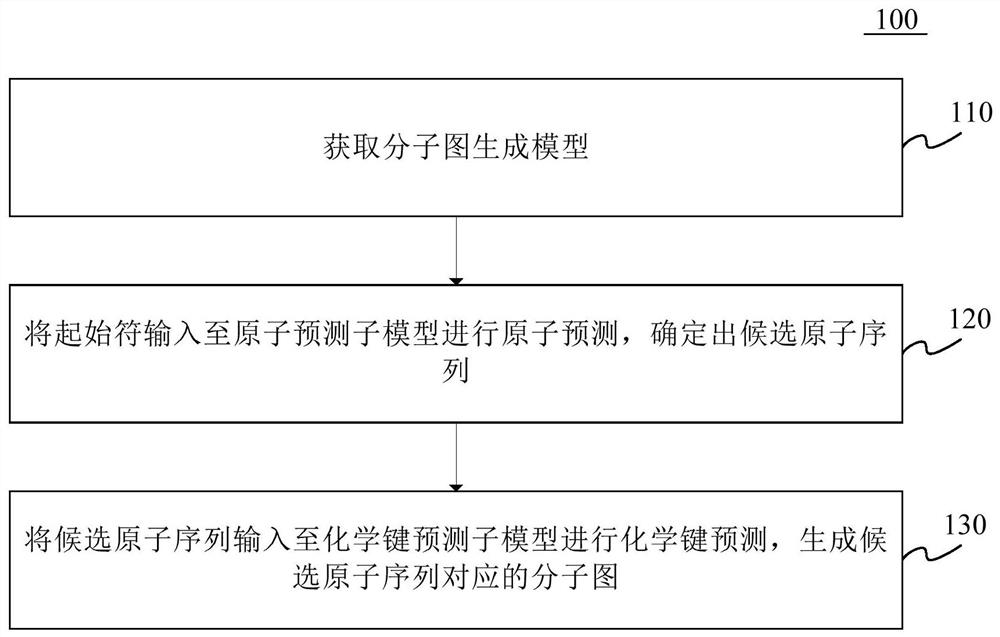
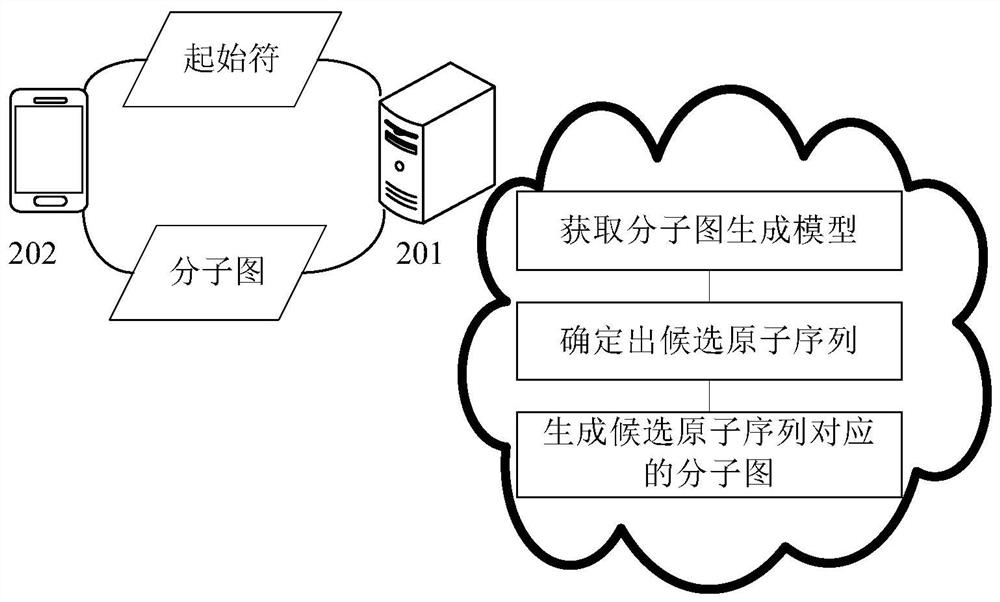
Molecular map generation method and device
PendingCN114822721AEasy to understandChemical property predictionMolecular designComputational physicsComputer science
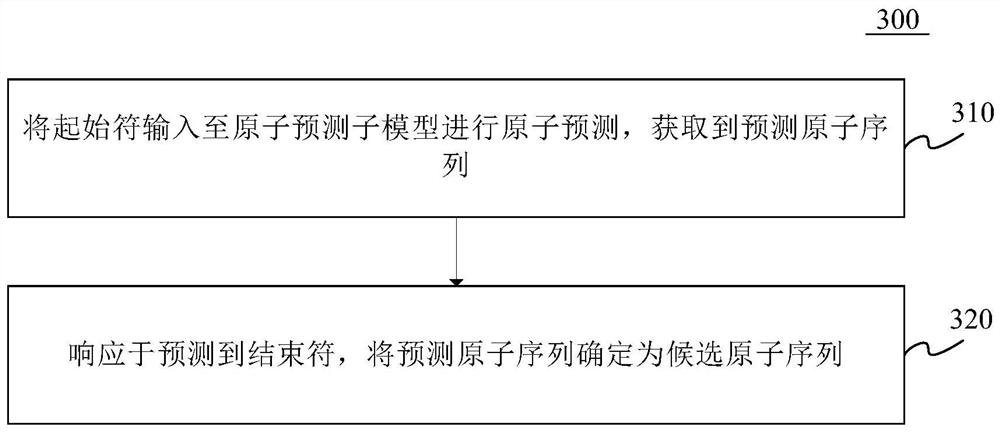
The invention provides a molecular map generation method and device, and relates to the technical field of computers, in particular to the technical field of biological computing and the technical field of artificial intelligence. According to the specific implementation scheme, firstly, a molecular graph generation model is obtained, the molecular graph generation model comprises an atom prediction sub-model and a chemical bond prediction sub-model, then a start character is input into the atom prediction sub-model for atom prediction, and a candidate atom sequence is determined; finally, the candidate atom sequence is input into the chemical bond prediction sub-model for chemical bond prediction, a molecular graph corresponding to the candidate atom sequence is generated, generation of the molecular graph is completed through the two steps of candidate atom sequence generation and chemical bond generation between candidate atoms, the molecular graph generation effect can be effectively improved, and the generation efficiency of the molecular graph is improved. The molecular map generation difficulty is reduced, and the operation efficiency is improved.
Owner:BEIJING BAIDU NETCOM SCI & TECH CO LTD
Protein Thermodynamic Analysis Method Based on Monte Carlo Simulation
ActiveCN103699816BEfficient analysisSpeed up searchSpecial data processing applicationsSimulation basedProtein thermodynamics
The invention relates to the technical fields of biological computing and Monte Carlo simulation, and particularly discloses a Monte Carlo simulation-based protein thermomechanical-analysis method, which comprises the following steps of A, determining a protein energy model; B, simulating and calculating the state density of a protein system. According to the method, the whole thermomechanical process of protein folding can be efficiently analyzed and researched to further explore and research a protein folding process.
Owner:SHENZHEN INST OF ADVANCED TECH
Bacterial cell 3-8 decoder and cell computer
The invention provides a bacterial cell 3-8 decoder and a cell computer. 8 cells from a first cell to an eighten cell are composed of a first characteristic signal promoter, a second characteristic signal promoter, a third characteristic signal promoter, a constitutive promoter, a promoter inhibition element, a promoter deinhibition element, an indication signal inhibition element and an indication signal deinhibition element. The decoder is a multi-cell computing component, decoding calculation is completed through activation and inhibition operation on the promoter and the indication signal,and the decoder has the accuracy, flexibility and usability of calculation. The bacterial cell 3-8 decoder can be used in related applications requiring a decoder such as a bio-computer, a biosensorand the like and has a wide range of applications.
Owner:MINZU UNIVERSITY OF CHINA
Bacterial Cell 3-8 Decoder and Cell Computer
The invention provides a bacterial cell 3-8 decoder and a cell computer. 8 cells from a first cell to an eighten cell are composed of a first characteristic signal promoter, a second characteristic signal promoter, a third characteristic signal promoter, a constitutive promoter, a promoter inhibition element, a promoter deinhibition element, an indication signal inhibition element and an indication signal deinhibition element. The decoder is a multi-cell computing component, decoding calculation is completed through activation and inhibition operation on the promoter and the indication signal,and the decoder has the accuracy, flexibility and usability of calculation. The bacterial cell 3-8 decoder can be used in related applications requiring a decoder such as a bio-computer, a biosensorand the like and has a wide range of applications.
Owner:MINZU UNIVERSITY OF CHINA
Method for bio-computer analysis for evaluating the risk for the onset of age-related macular degeneration
InactiveUS20190233896A1Reliable and definitely more reproducible resultEstimate the genetic riskHealth-index calculationMicrobiological testing/measurementGenetic riskRAD51
The present invention relates to a method for estimating the risk factor (HR) for the onset of macular degeneration in a subject, which method comprises the evaluation of a first series of genetic parameters comprising at least the genetic loci R1210C in CFH, variants in COL8A1 and RAD51 B, and the evaluation of a second series of environmental, individual and / or clinical parameters. This method is characterized in that said risk factor (HR) is calculated by assigning a greater importance to said evaluation of the first series of genetic parameters with respect to said evaluation of the second series of environmental, individual and / or clinical parameters. The method of the present invention thus allows to estimate the genetic risk of AMD more accurately than the methods known in the art.
Owner:S I F I SPA
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com