A method for analyzing the geometric structure characteristics of DNA bin protein interface
A technology for binding proteins and analysis methods, which is applied in the field of analysis of the geometric structural characteristics of DNA-binding protein interfaces, and can solve the problems of incomplete elucidation of DNA repair, transcription, recombination, and lack of structural mechanisms.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0033] The specific embodiments of the present invention will be further described below in conjunction with the drawings.
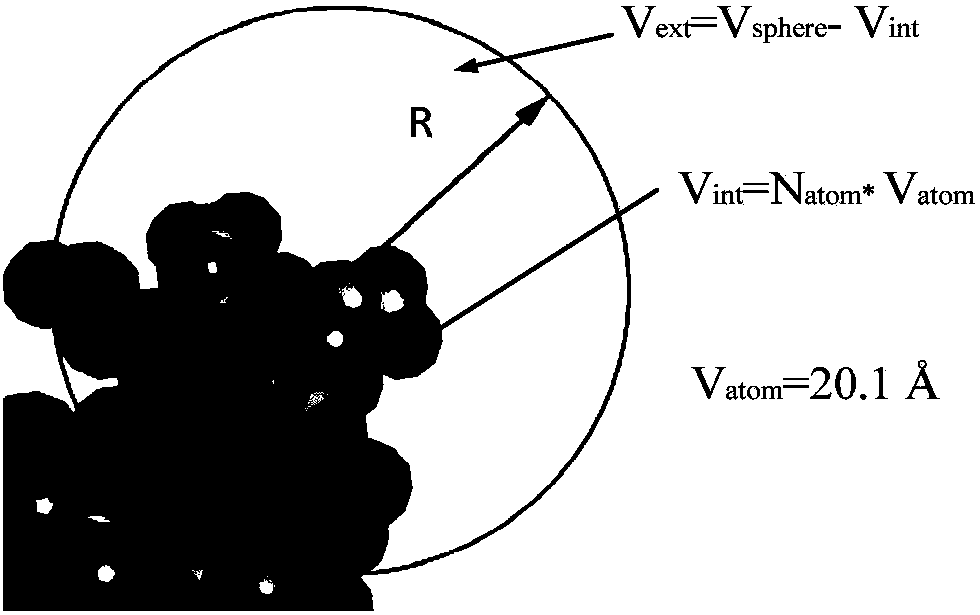
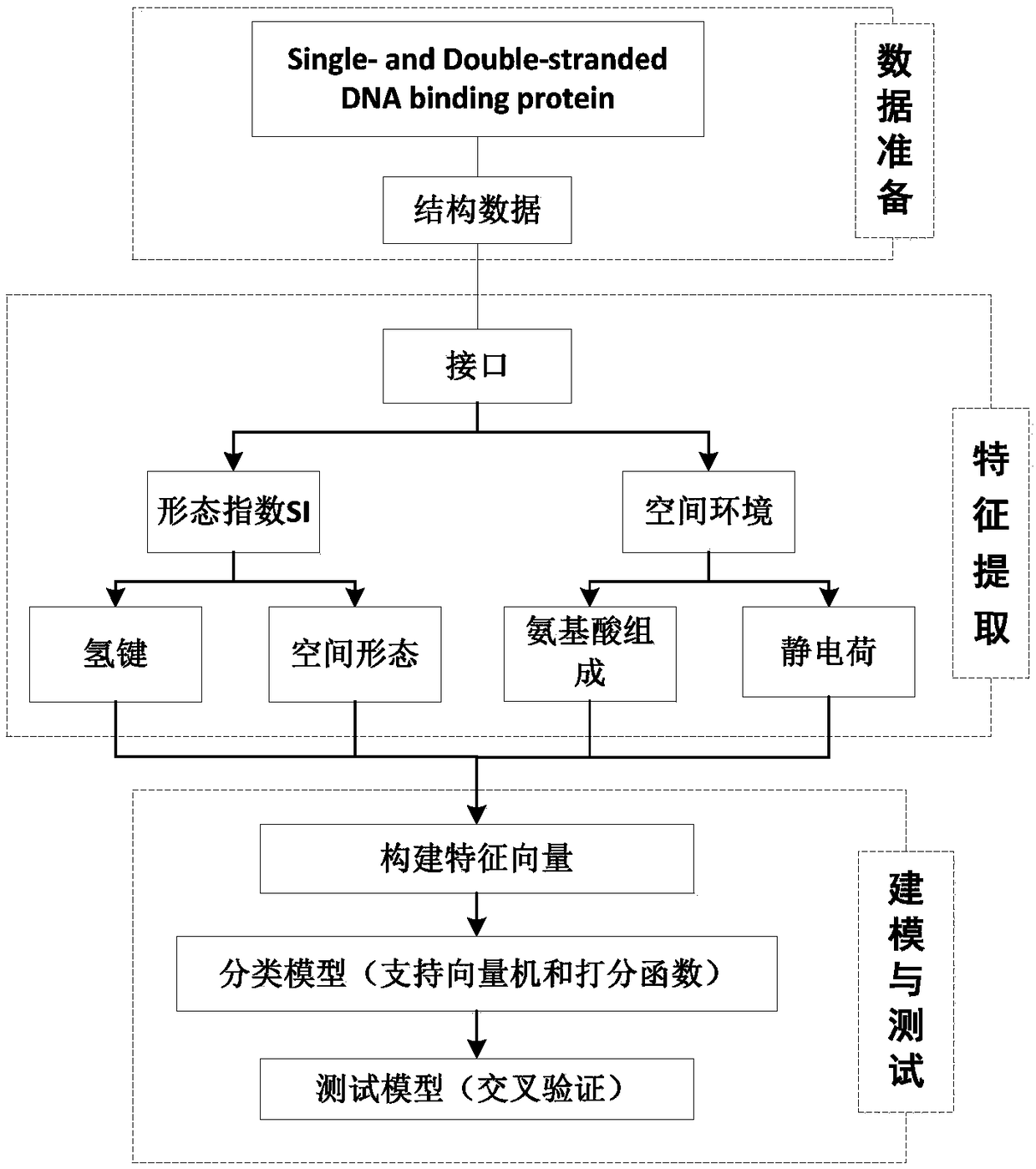
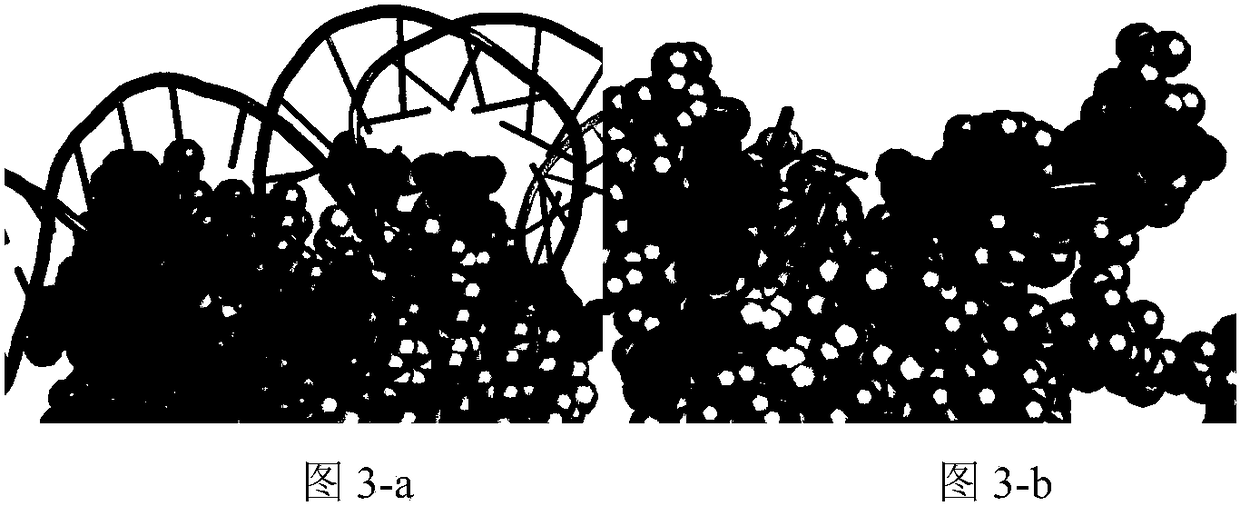
[0034] The present invention extracts protein-DNA binding sites from a protein-DNA composite data set, and extracts structural features of the binding sites according to the binding sites, which are divided into two types of features: morphological index SI features and spatial environment features. On the basis of the morphological index characteristics, the hydrogen bond distribution of the interface and the frequency of the spatial shape are calculated; on the spatial environment characteristics, the composition distribution of amino acid residues and the electrostatic charge distribution characteristics are calculated. Construct a feature vector, and use SVM to build a classification model, and test the distinguishing performance of features. The process is as figure 2 As shown, the specific implementation steps are as follows:
[0035] 1. Extract the p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com