Patents
Literature
45 results about "Protein dna" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
DNA is NOT a protein. DNA is made up from a chain of nucleotides. Proteins are built from a long chain of amino acids. Apart from the fact they're chaining together separate smaller molecules, they have very little in common. 3 nucleotides in DNA translate to one specific amino acid and there is some duplication.
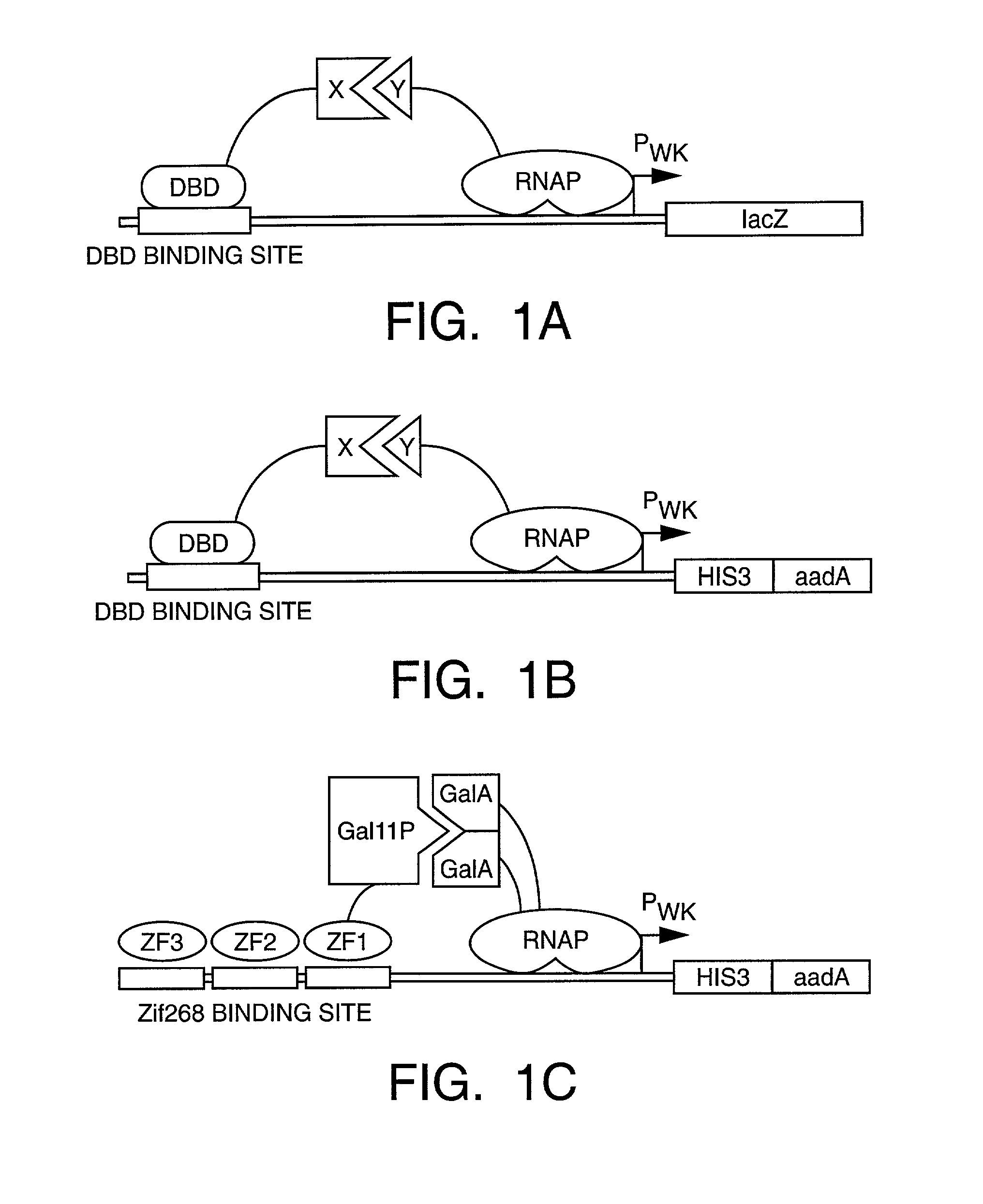
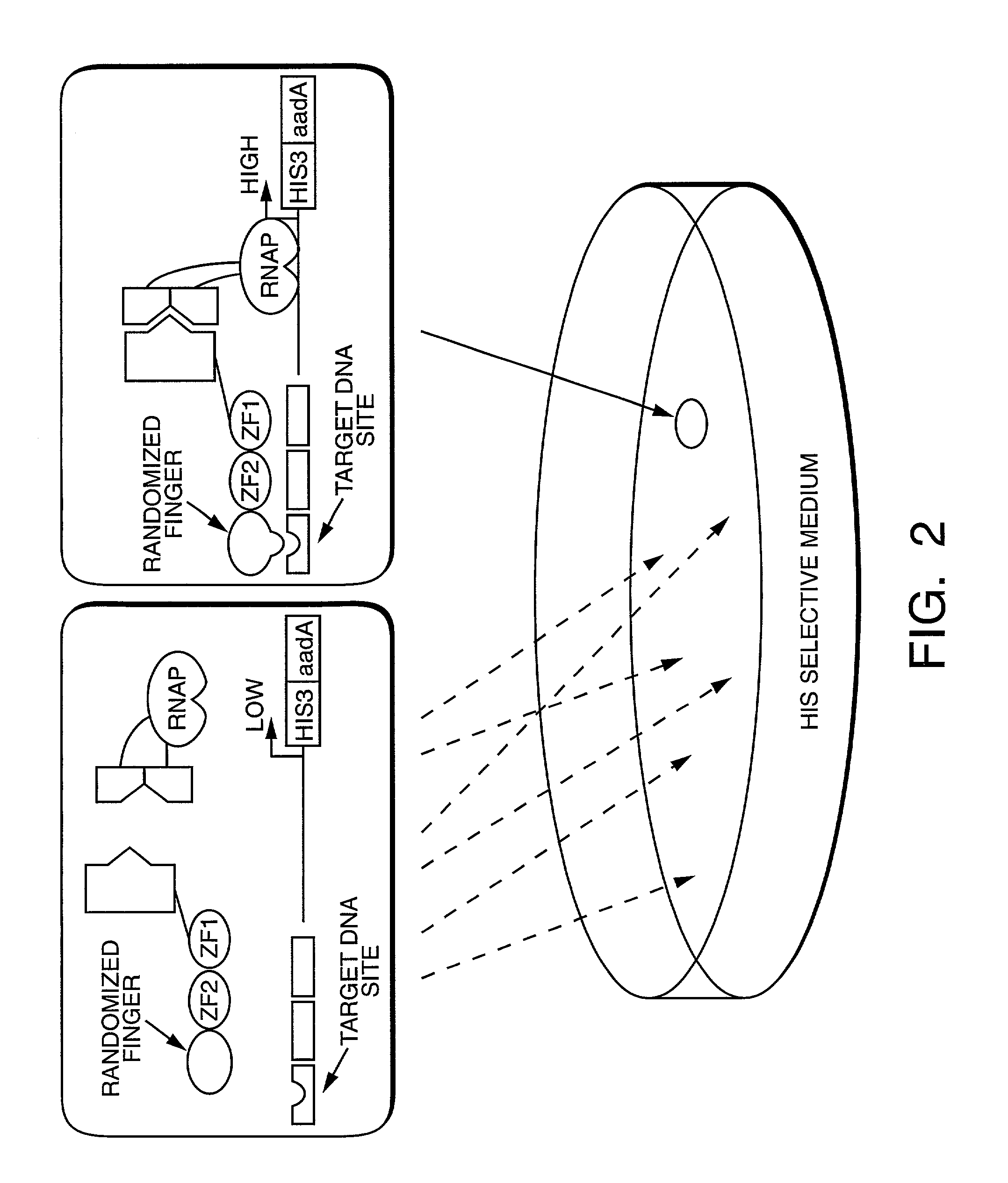
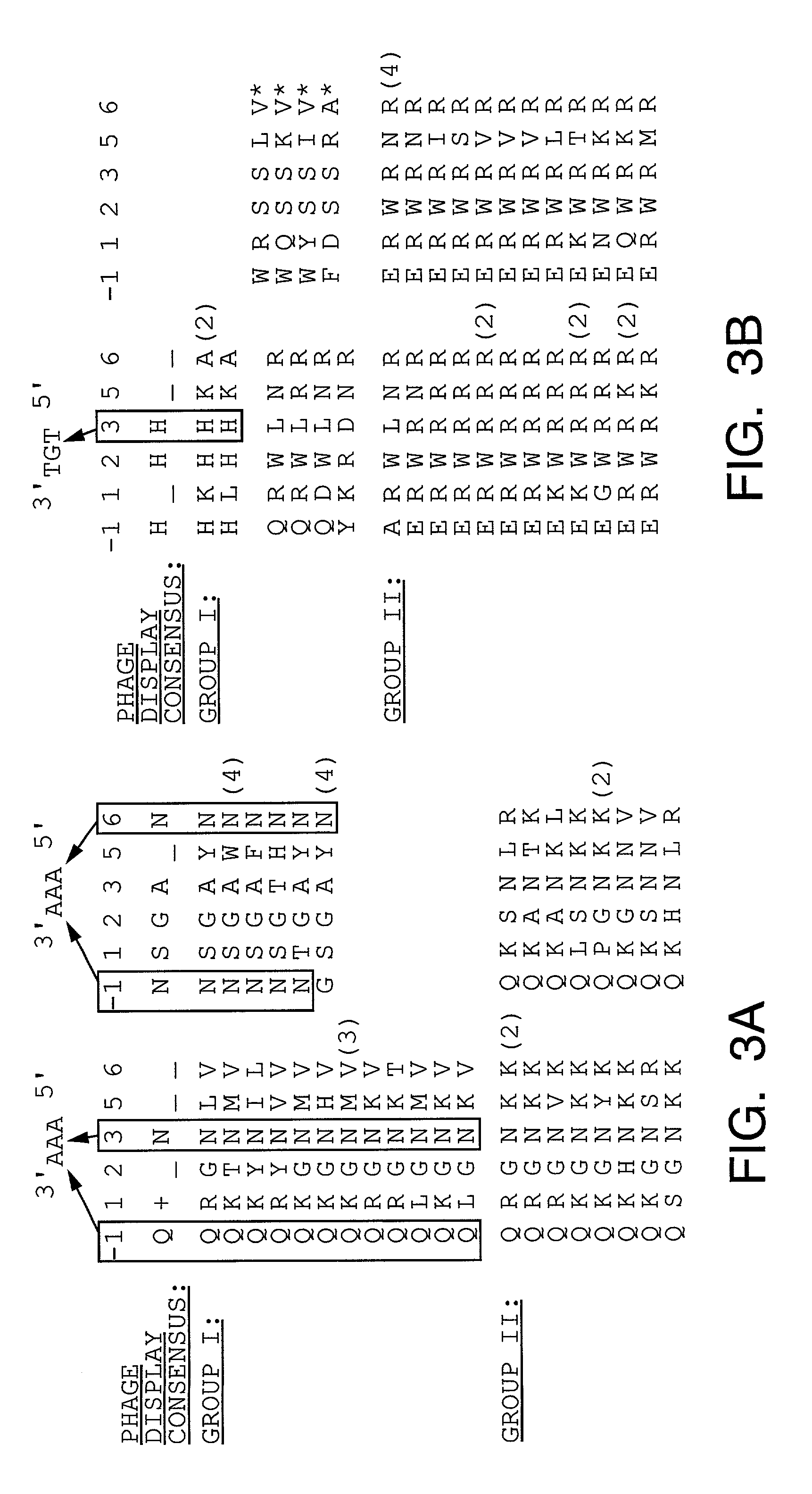
Methods and compositions for interaction trap assays
InactiveUS7029847B2Increase opportunitiesEnhanced interactionMicrobiological testing/measurementBiological testingProtein targetNucleic acid sequencing
The present invention provides methods and compositions for interaction trap assays for detecting protein-protein, protein-DNA, or protein-RNA interactions. The methods and compositions of the invention may also be used to identify agents which may agonize or antagonize a protein-protein, protein-DNA, or protein-RNA interaction. In certain embodiments, the interaction trap system of the invention is useful for screening libraries with greater than 107 members. In other embodiments, the interaction trap system of the invention is used in conjunction with flow cytometry. The invention further provides a means for simultaneously screening a target protein or nucleic acid sequence for the ability to interact with two or more test proteins or nucleic acids.
Owner:MASSACHUSETTS INST OF TECH
Magnetic fluorescent double-function nanoion probe and preparation method thereof
InactiveCN104745192AEasy to prepareEasy to manufactureMaterial nanotechnologyNanomagnetismMagnetite NanoparticlesComposite nanoparticles
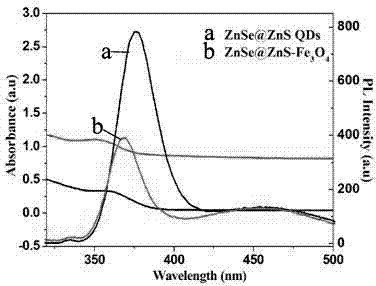
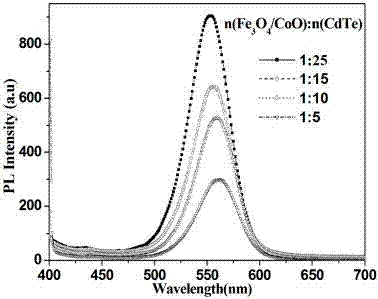
The invention belongs to the field of preparing a nanomaterial, and particularly relates to a magnetic fluorescent double-function nanoion probe and a preparation method thereof. The composite microparticles have the fluorescent performance of quantum dots and the magnetism performance of magnetic nanoparticles, and can be used as target positioning in biological body and biological fluorescent imaging aspects. The invention provides a preparation method of the magnetic fluorescent double-function nanomaterial, and the magnetic fluorescent double-function nanomaterial is obtained by taking chitosan-modified magnetic nanoparticles as a core and connecting water-soluble quantum dots through adopting an ionic crosslinked method, the fluorescent quantum dots are distributed on the surfaces of the magnetic nanoparticles with the particle size of 10-200nm, and the particle size of the quantum dots is 1.5-10nm. The preparation method is mild in reaction conditions, the operation method is simple, the prepared composite nanoparticles have good luminescence performance and magnetic performance, and have wide application prospects in the fields of biomarking, fluorescent immunoassay, bioseparation, protein DNA enrichment and separation, the preparation of drug carrying system, and target imaging and the like.
Owner:UNIV OF JINAN
Compositions and methods for identifying and testing TGF- beta pathway agonists and antagonists
InactiveUS6046165ASimple and sensitiveEasy to separateBiocidePeptide/protein ingredientsADAMTS ProteinsAgonist
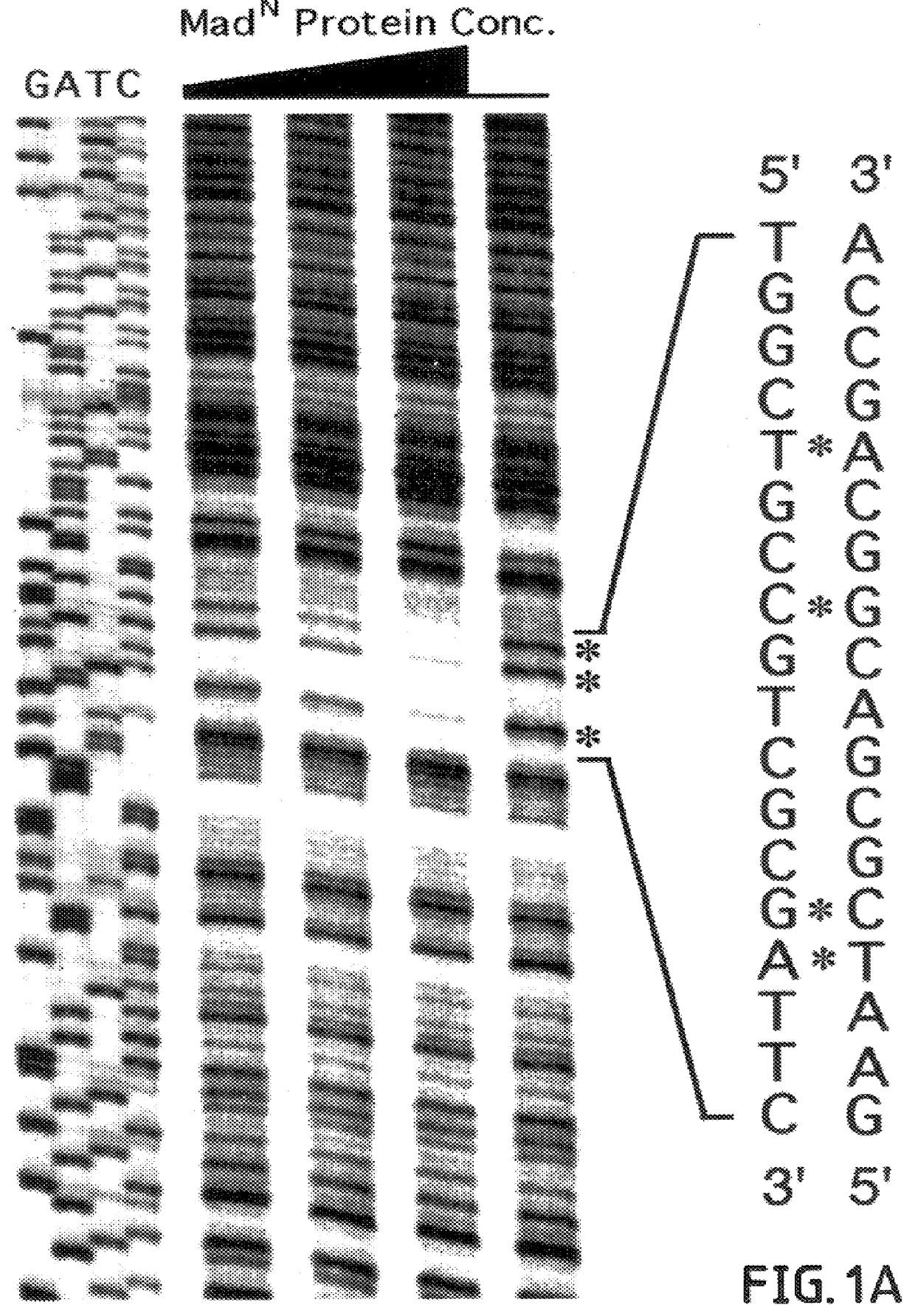
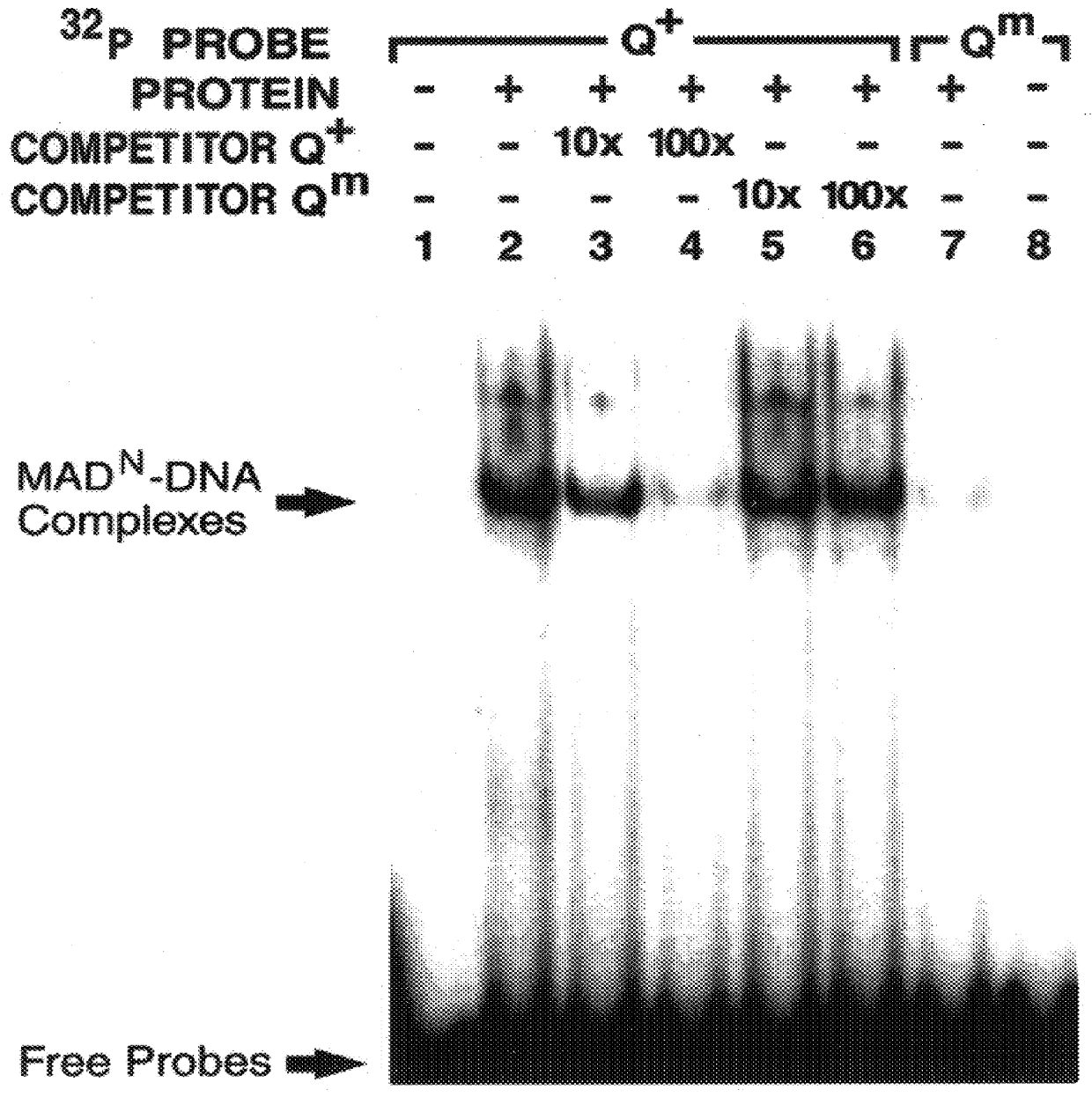
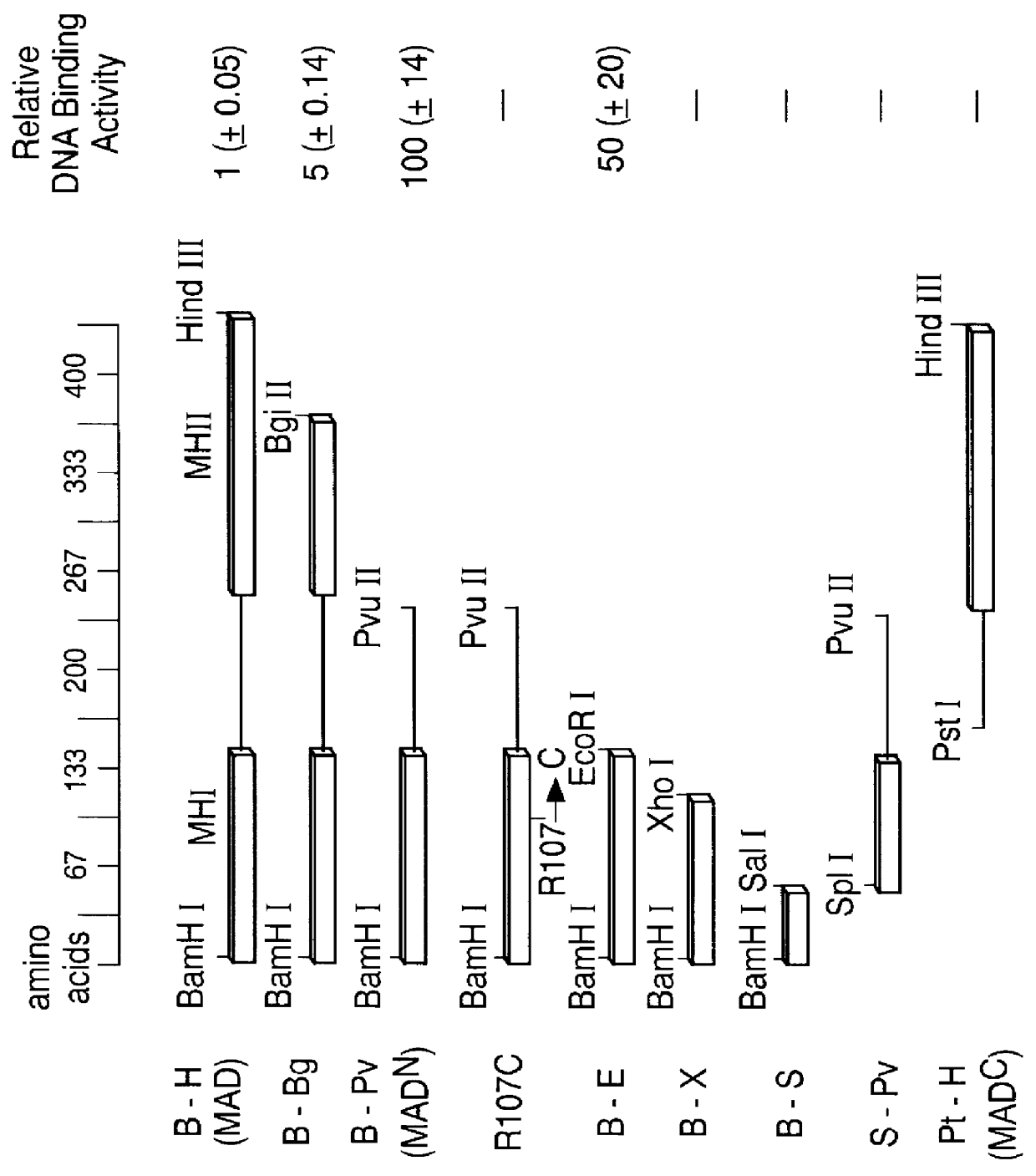
The invention provides compositions and methods of identifying and testing TGF- beta pathway agonists and antagonists, and in particular compositions comprising Mothers against DPP (MAD) proteins and related Smad polypeptides which exhibit sequence-specific DNA-binding activity. The invention also provides a novel DNA sequence (SEQ ID NO:19); (SEQ ID NO:20); (SEQ ID NO:21) that is bound with high affinity by Drosophila MAD protein. This protein is useful for identifying compounds that will enhance or interfere with MAD protein-DNA binding.
Owner:WISCONSIN ALUMNI RES FOUND +1
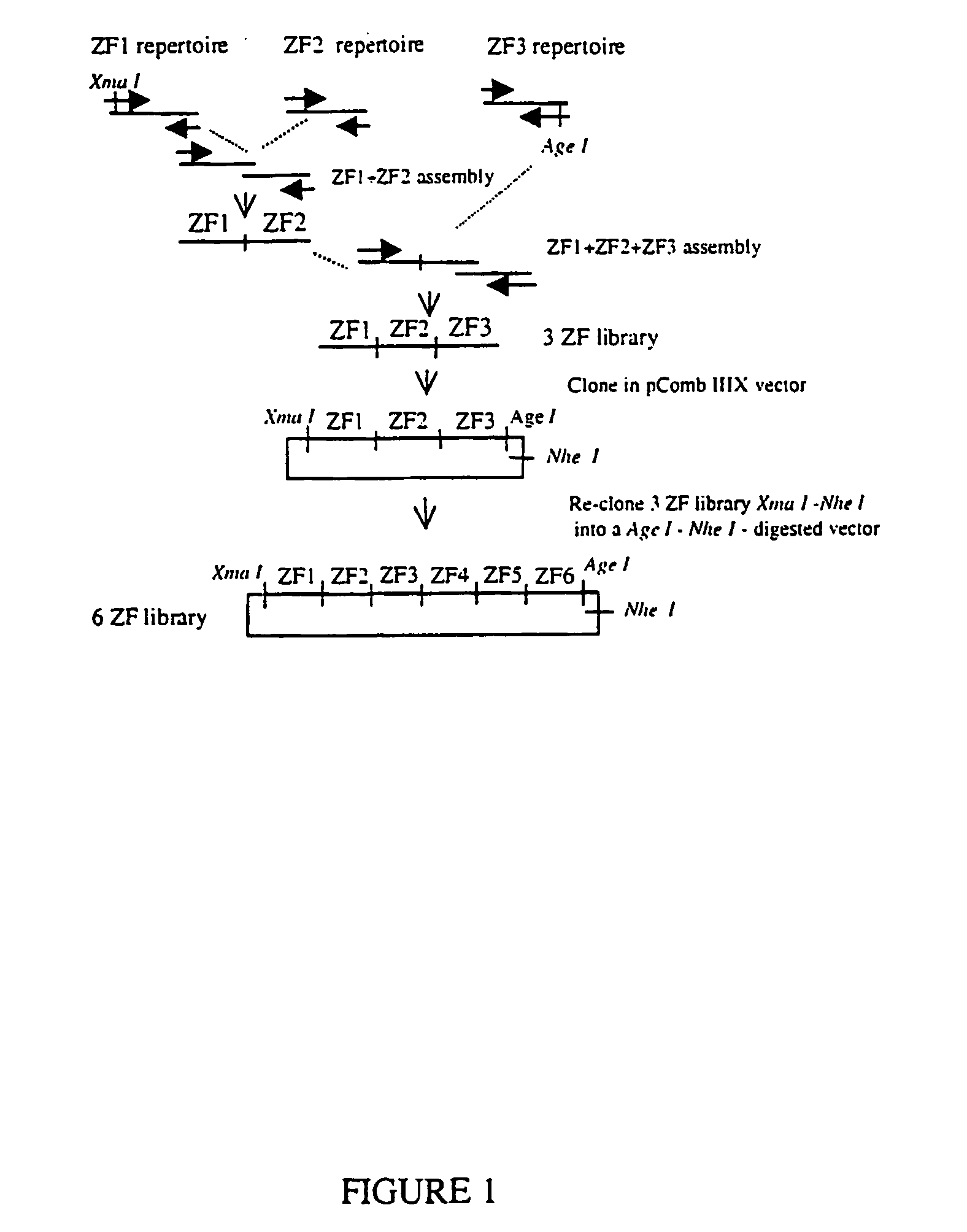
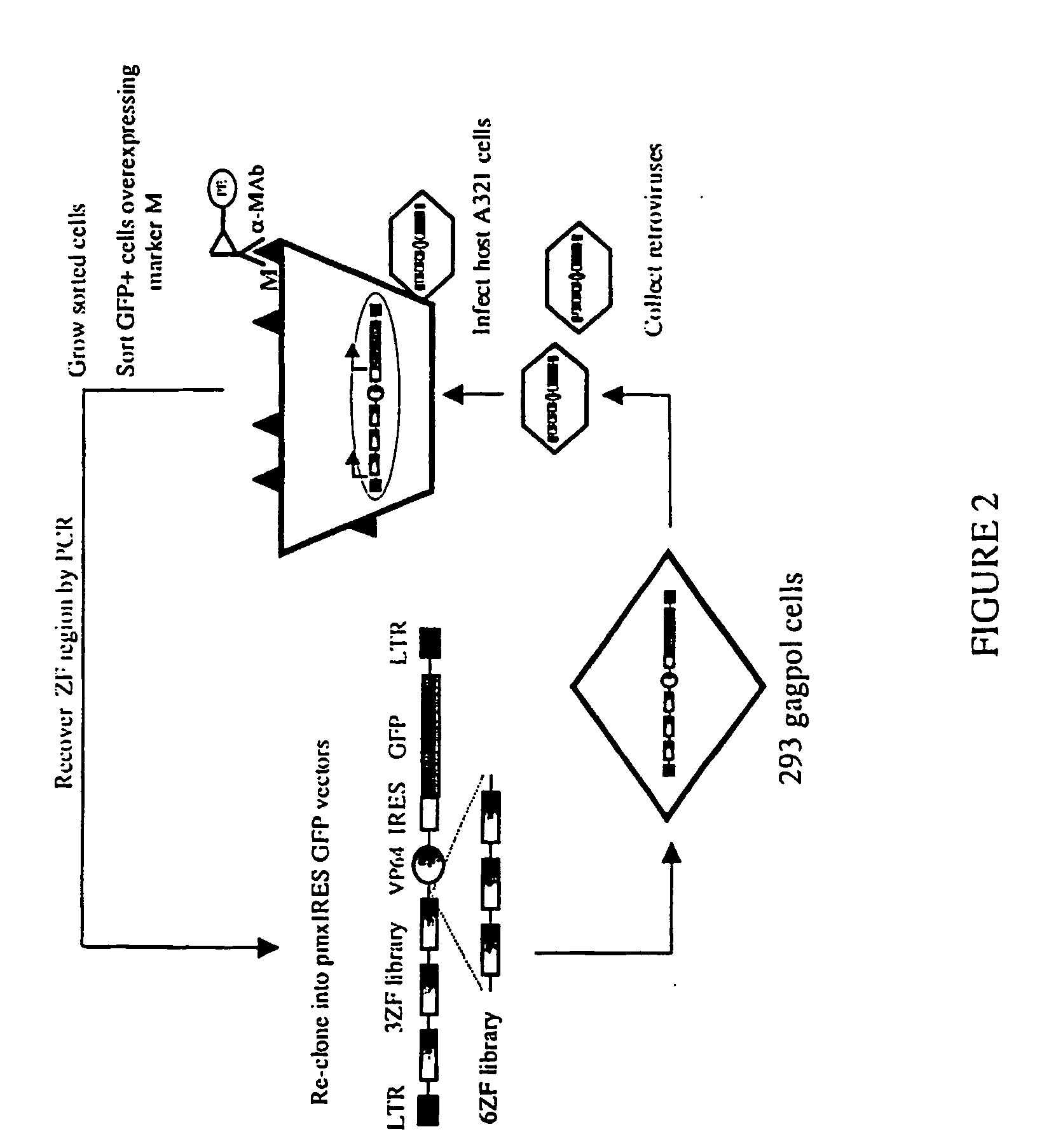
Zinc finger libraries
A library of multimeric DNA binding polypeptides is provided. Preferred such polypeptides are zinc finger protein DNA binding domains. Libraries of nucleotides encoding such polypeptides, expression vectors containing such nucleotides, cells containing any of the libraries and uses of the libraries are also provided.
Owner:THE SCRIPPS RES INST
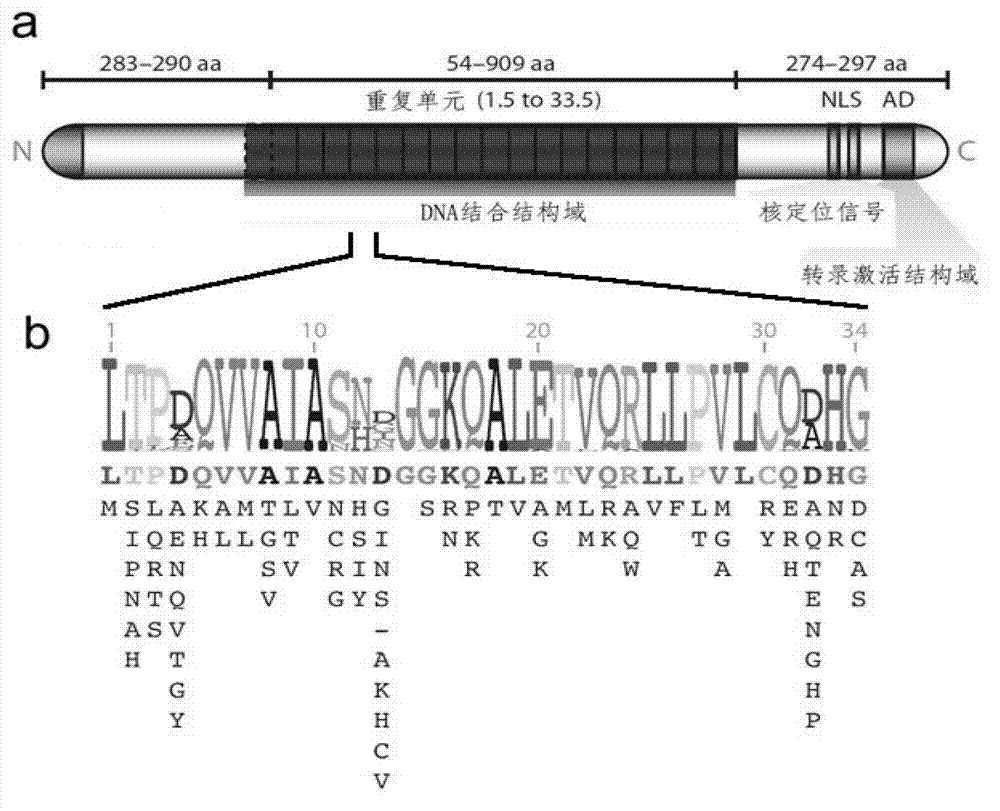
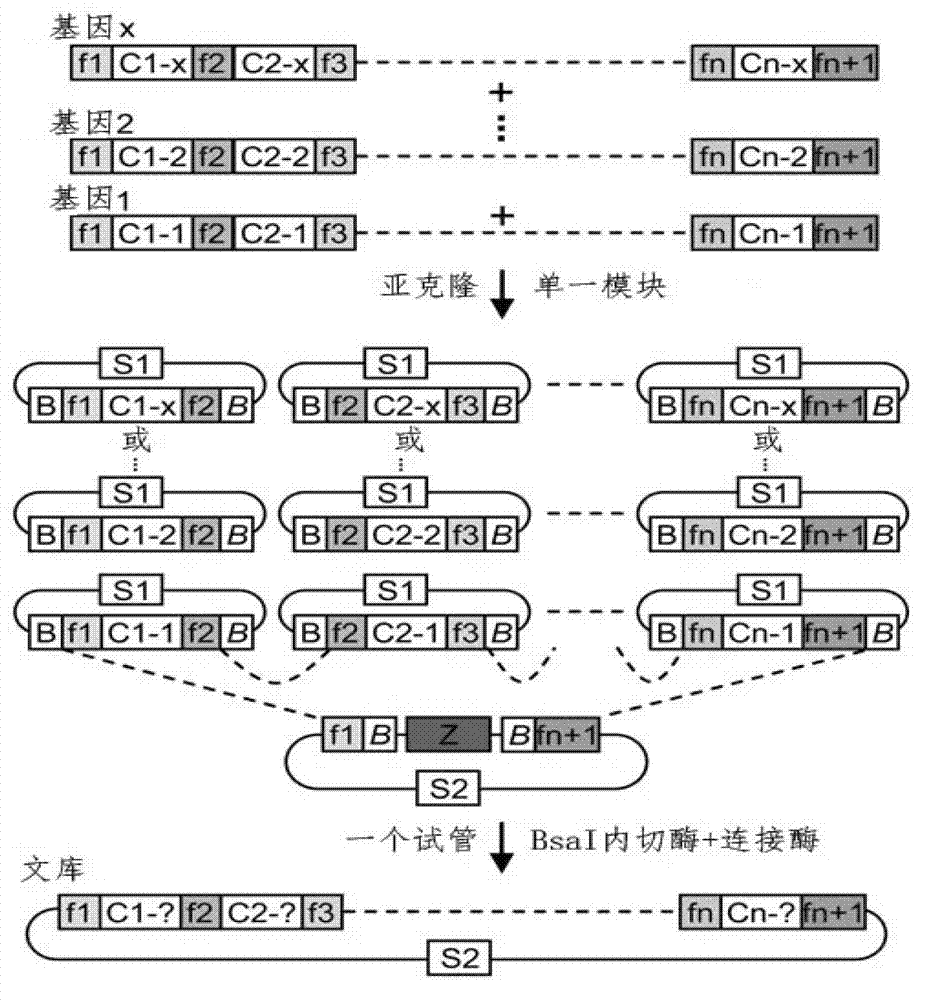
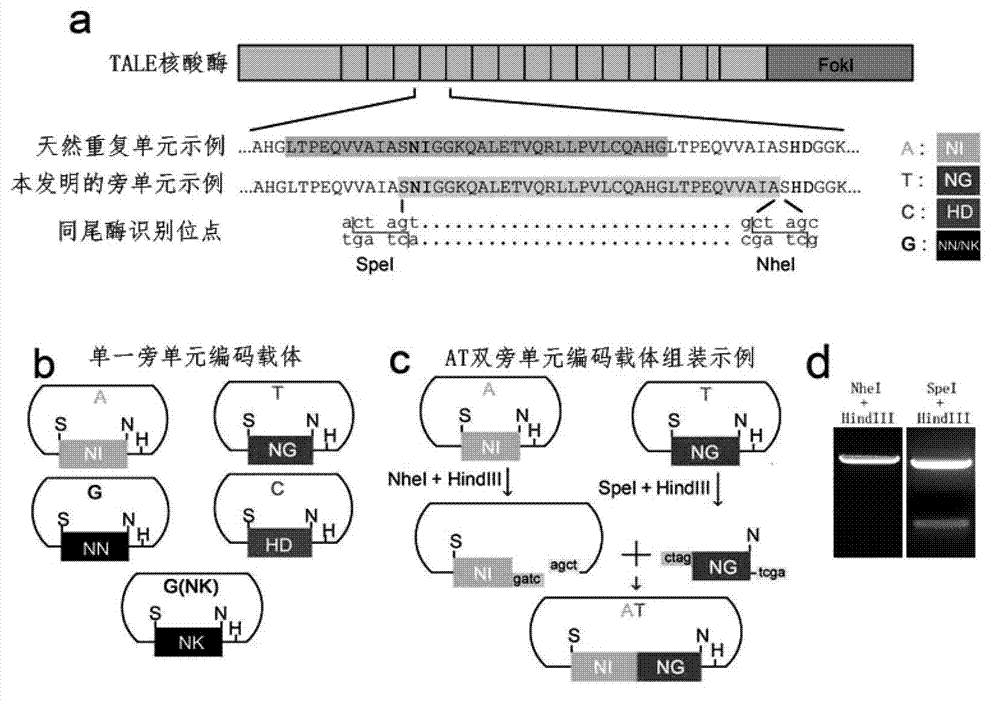
Method for building TALE (transcription activator-like effector) repeated sequences
ActiveCN102787125AHas a cumulative effectShorten the timeMicroorganism based processesFermentationNucleotideTAL effector
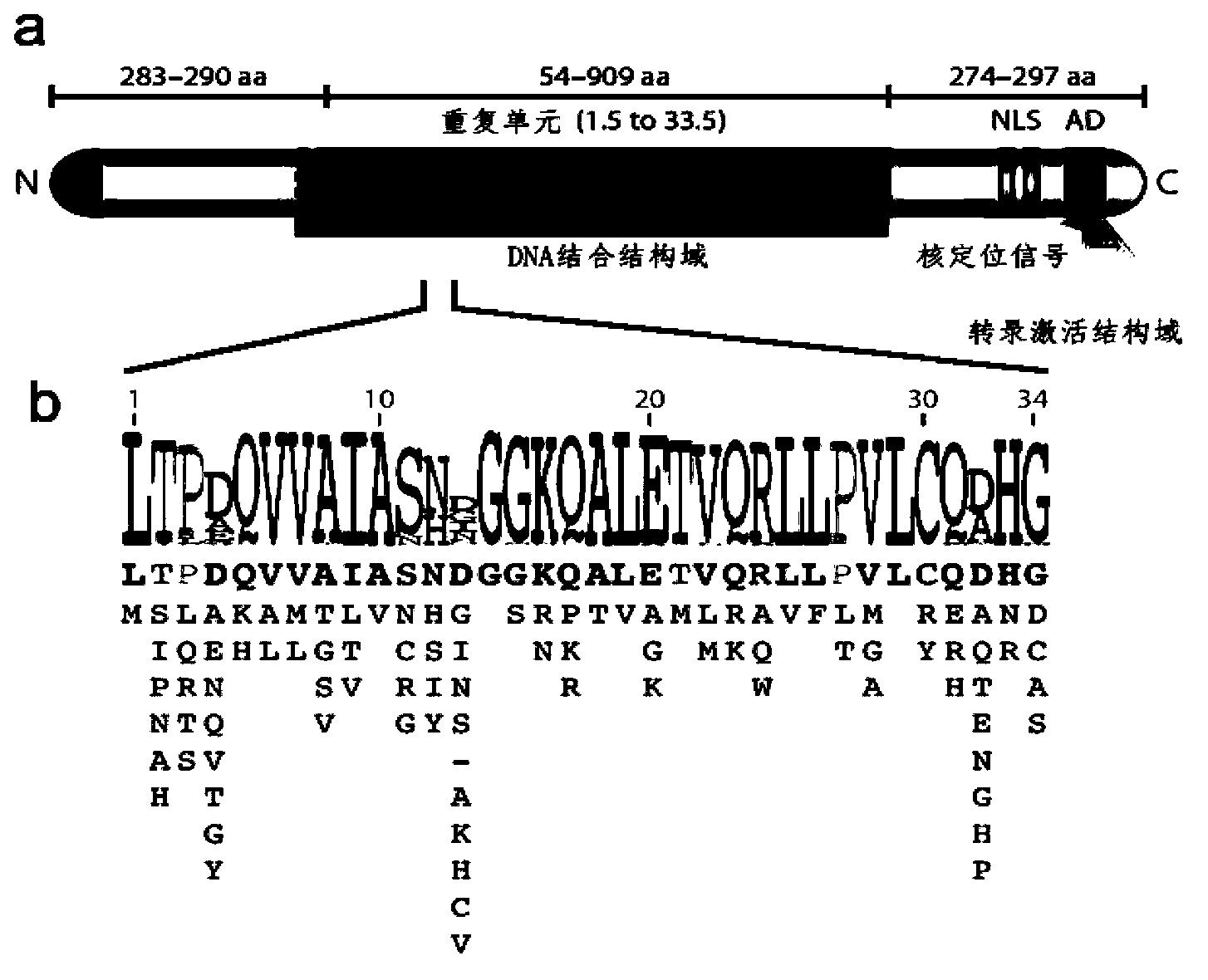
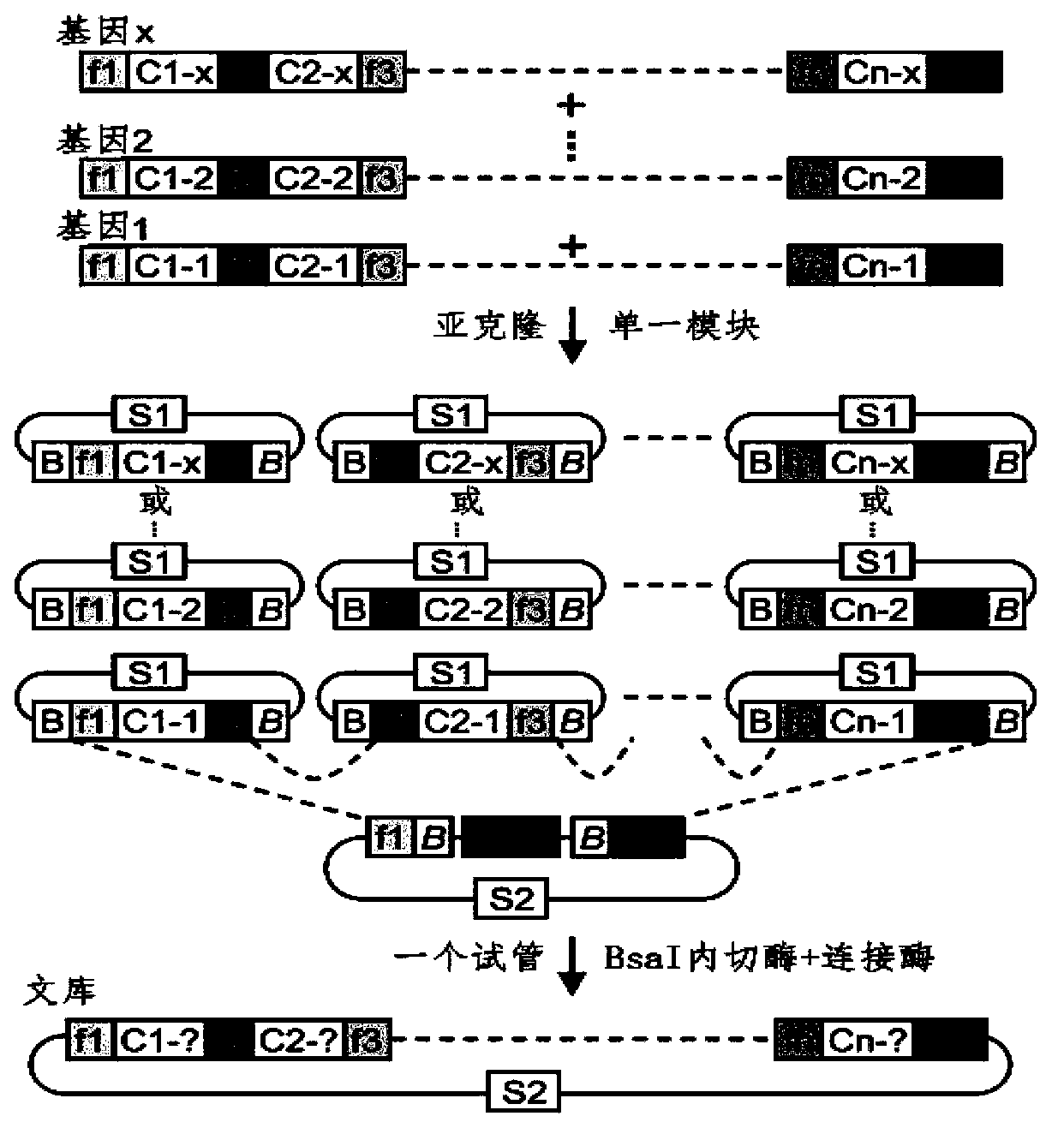
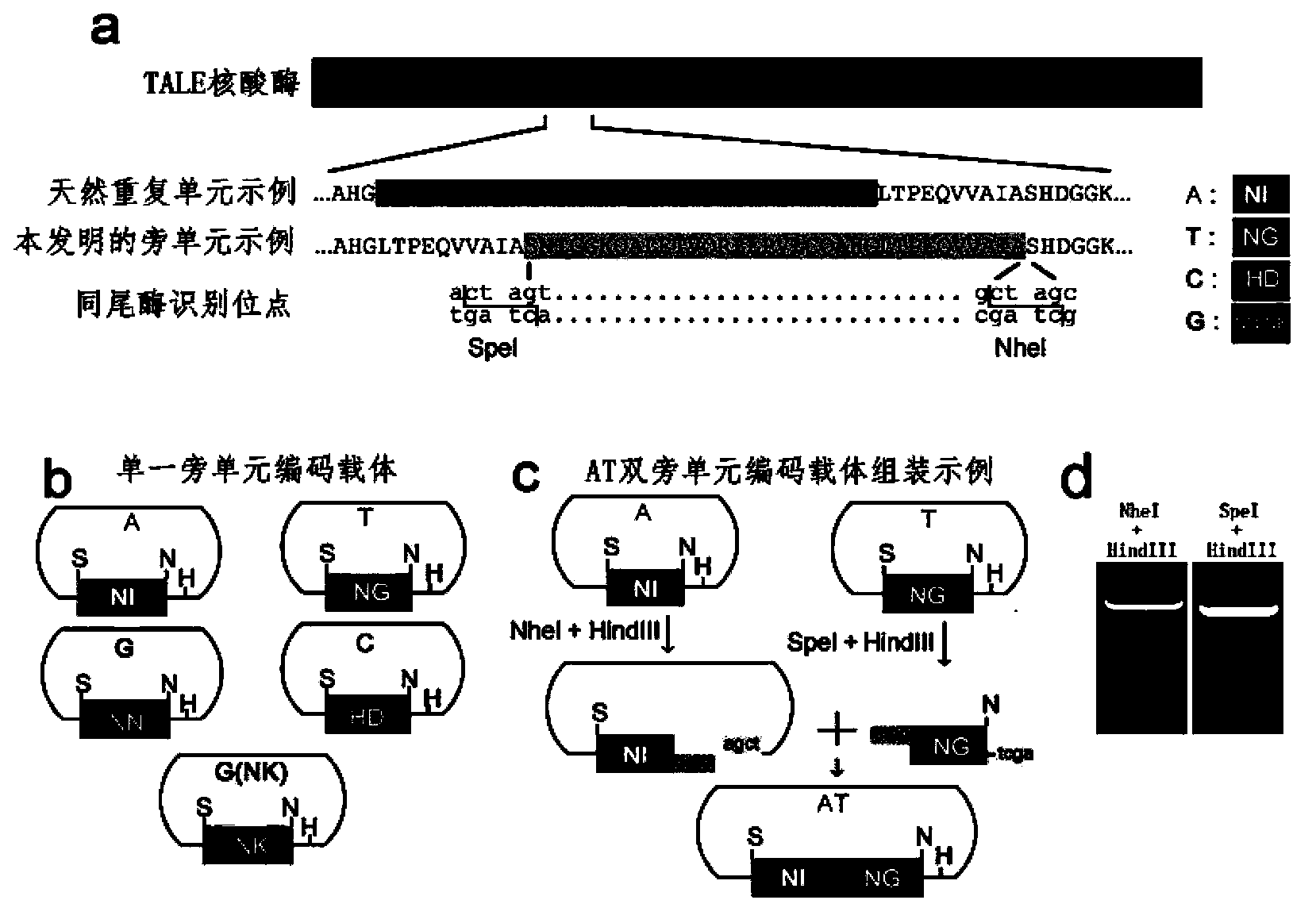
The invention discloses a side unit used for building TALE (transcription activator-like effector) repeated sequences. The side unit is a repeated unit DNA (deoxyribonucleic acid) segment containing isocaudarner or different flat terminase identification sites at two ends, and the preated unit DNA segment codes repeated units of RVD (repeated variable di-residue) containing NI, NG, HD, NK or NN or variants of the repeated units, wherein in the 5'end isocaudarner or flat terminase identification sites, the 3' end of the identification sites at least has one nucleotide participating in the amino acid coding the N end of the side unit; and in the 3' end isocaudarner or flat terminase identification sites, the 5' end of the identification sites at least has one nucleotide participating in the amino acid coding the C end of the side unit. The side unit has the advantages that the TALE repeated sequences containing any repeated unit number and any ranging sequences, plasmid vectors containing TALE repeated sequences, coding TALE protein DNA combined structural domains and various derived fusion protein plasmid vectors can be conveniently built.
Owner:PEKING UNIV
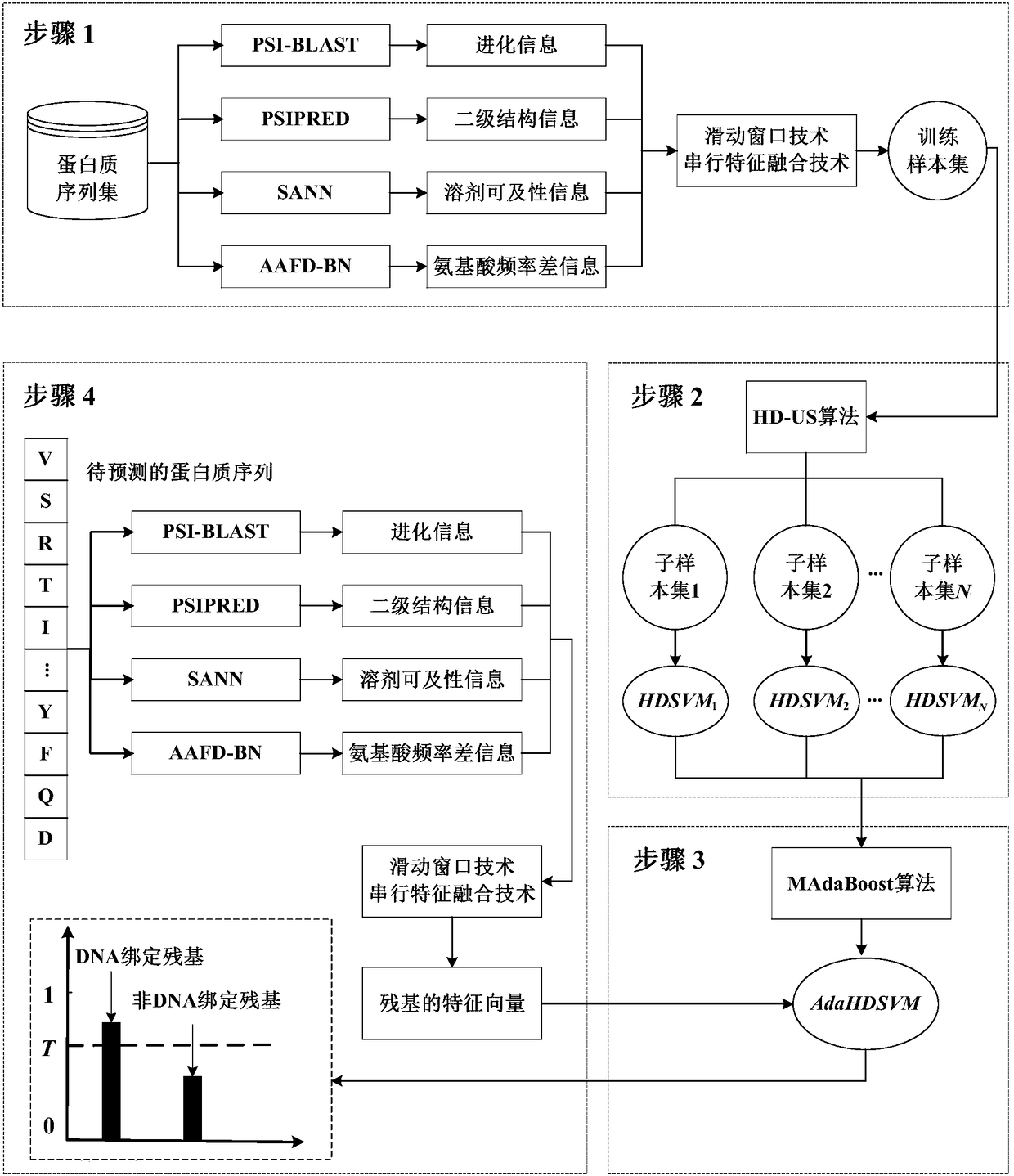
Protein-DNA binding residue prediction method based on sampling and integrated learning
InactiveCN109147866AImprove forecast accuracyReduce lossProteomicsGenomicsFeature extractionAlgorithm
The invention discloses a protein-DNA binding residue prediction method based on sampling and integrated learning. The method comprises the steps of (1) feature extraction and training sample set construction, (2) sampling and model training, (3) model integration, and (4) online prediction. The method is used for solving the shortcomings of low prediction precision caused by the problems of few feature types and class imbalance in protein-DNA binding residue prediction problems and has the advantages of high prediction precision and high generalization ability.
Owner:NANJING UNIV OF SCI & TECH
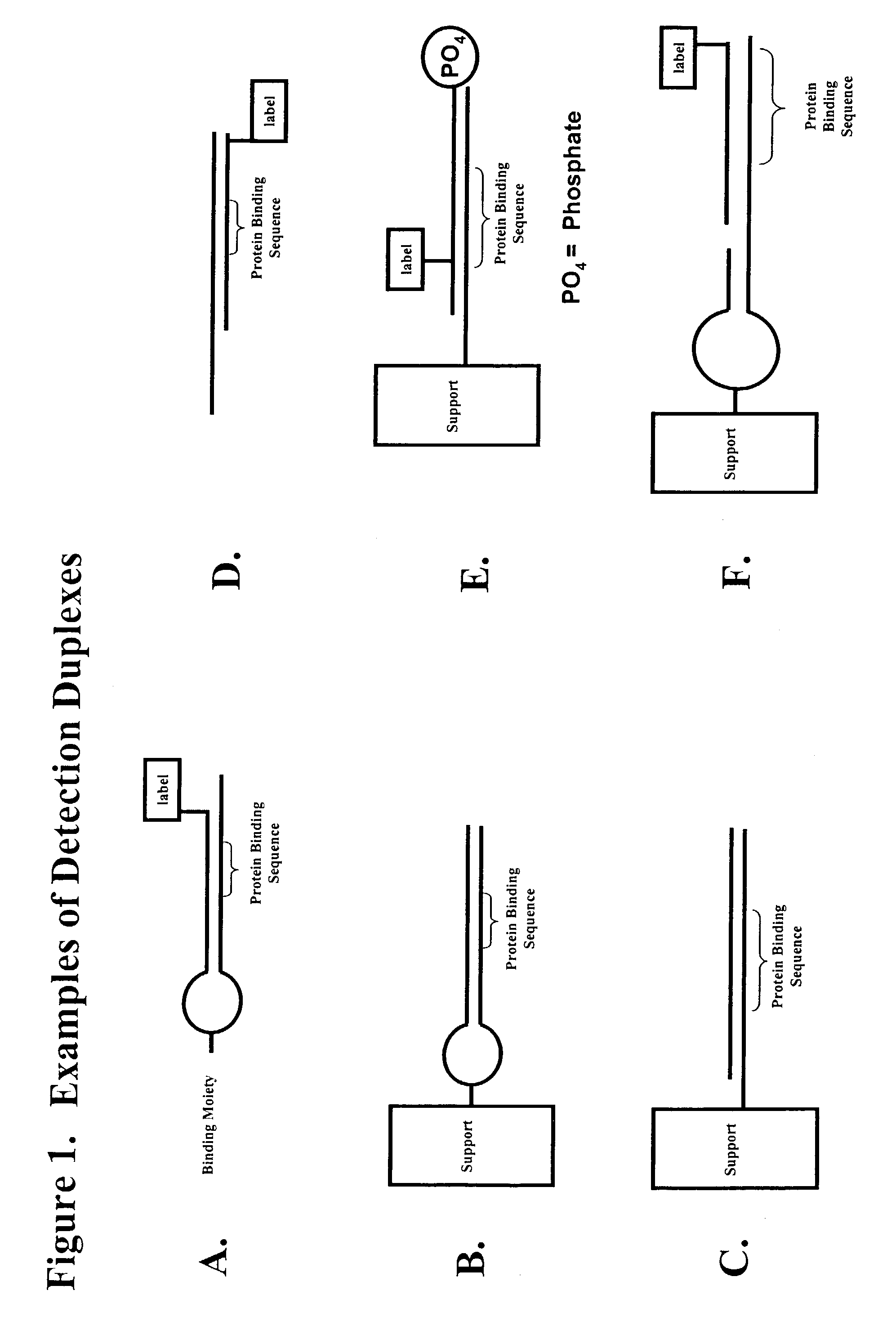
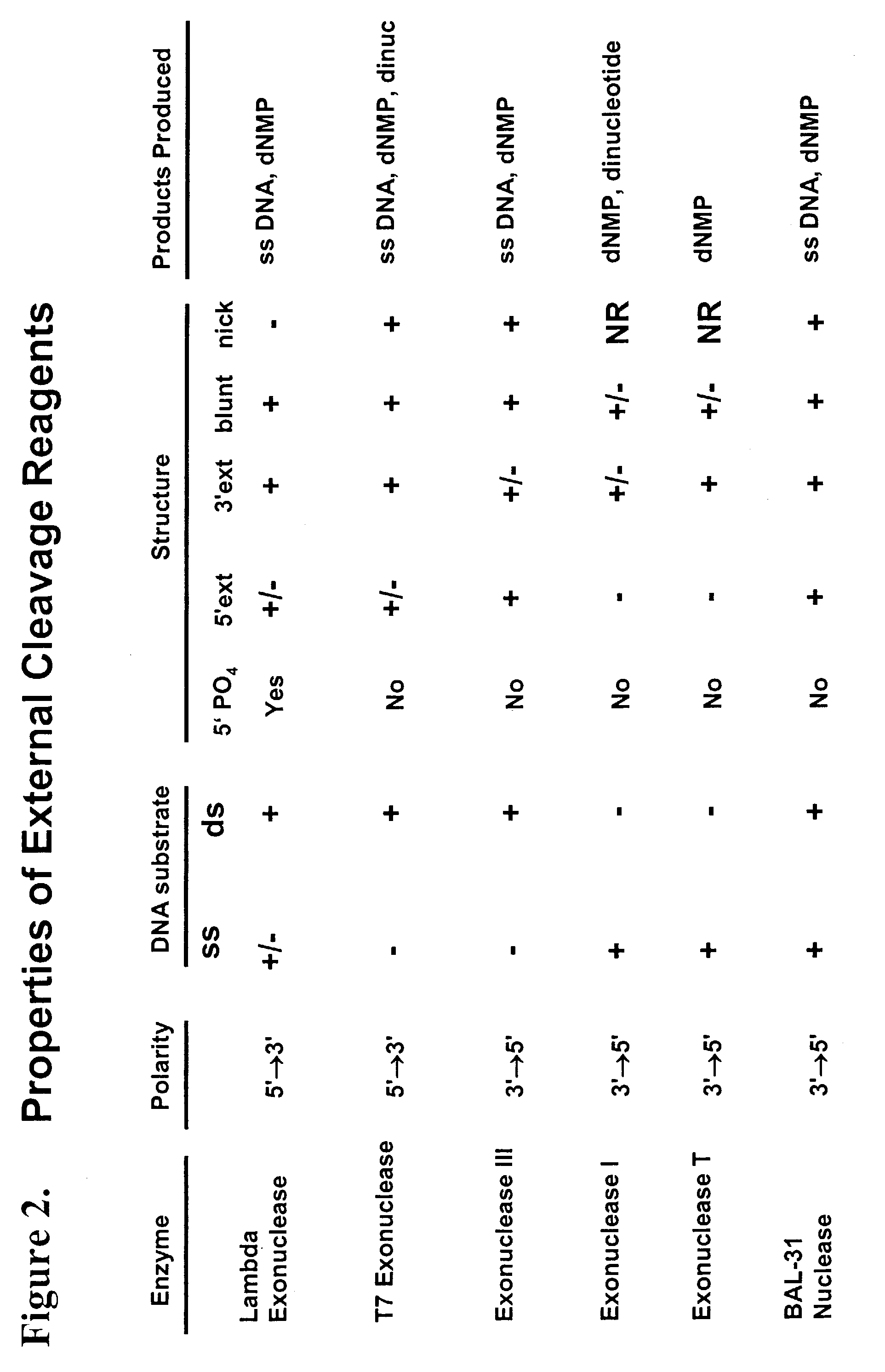
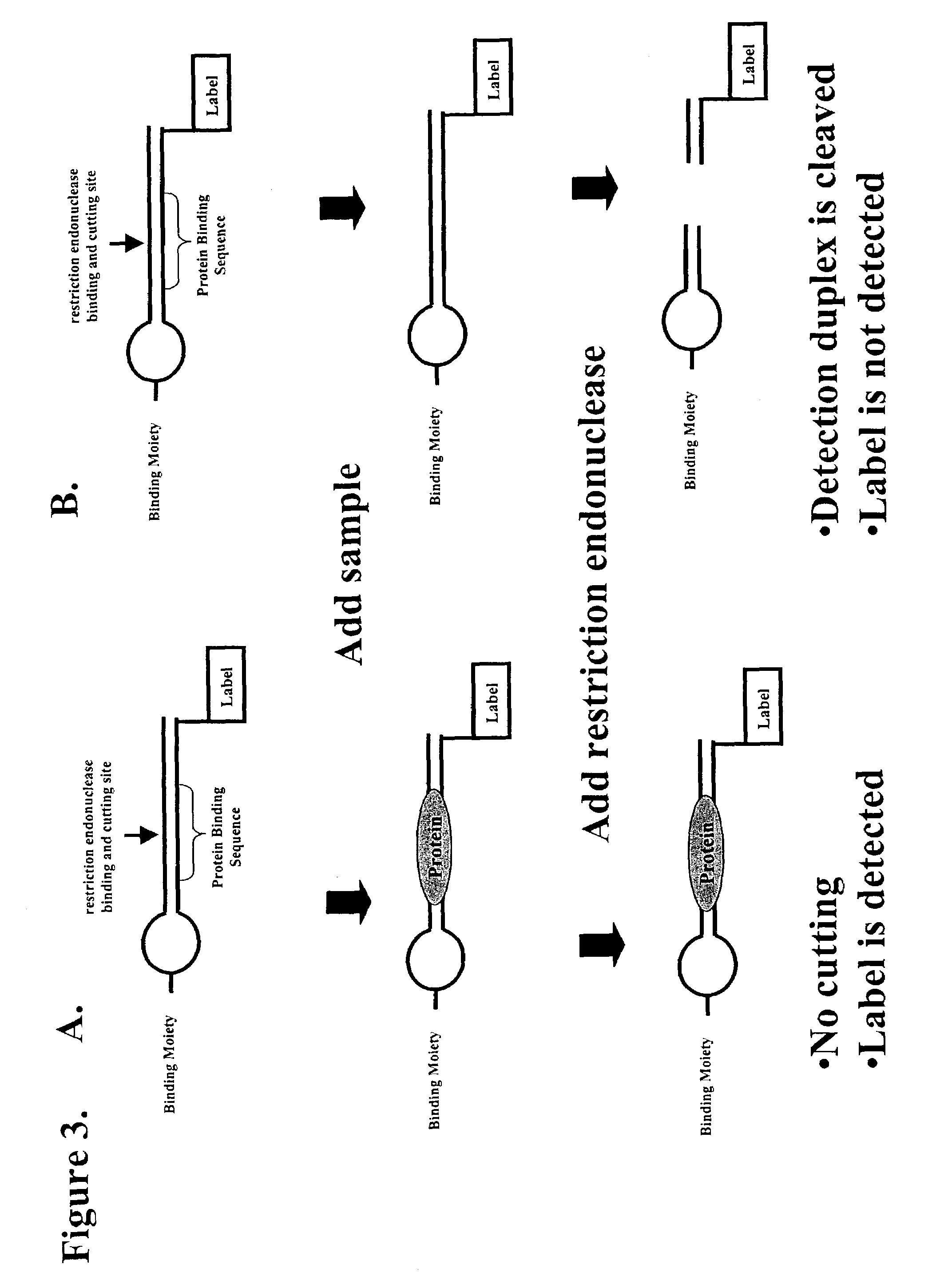
Multiplex method of detecting sequence-specific DNA binding proteins using detection duplexes comprising unmodified nucleic acid sequences as capture tags
InactiveUS7122317B2Microbiological testing/measurementBiological material analysisSingle sampleSequence-Specific DNA Binding Protein
Compositions and methods are provided for detecting and measuring DNA-binding proteins. The compositions and methods permit the simultaneous or near-simultaneous detection of multiple DNA-binding proteins in a multiplex or array format, and can be used to generate profiles of DNA binding activity by proteins, specifically, transcription factors. Multiple protein-DNA binding events in a single sample may be detected and quantitated in a high-throughput format.
Owner:MARLIGEN BIOSCI
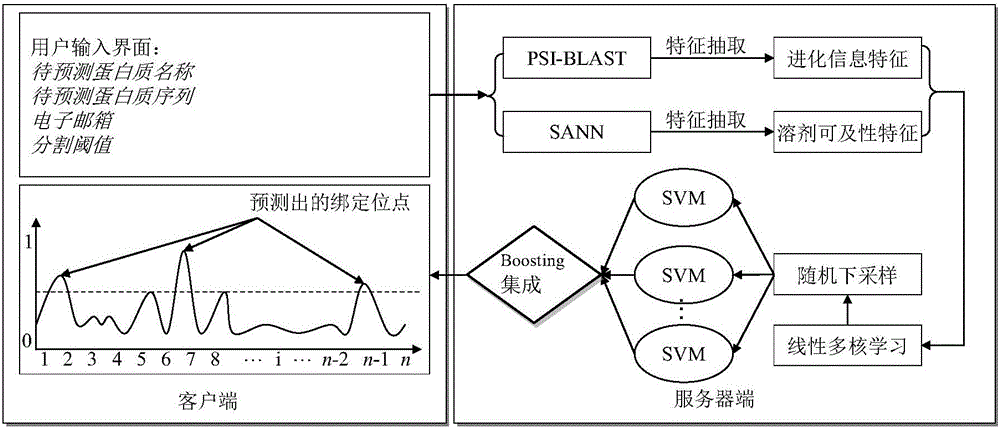
Multi-core-learning and Boosting algorithm based protein-DNA binding site prediction method
InactiveCN105808975AImprove forecast accuracyImprove interpretabilityProteomicsGenomicsSolvent accessibilityAlgorithm
The present invention provides a multi-core-learning and Boosting algorithm based protein-DNA binding site prediction method. The method comprises the following steps: feature extraction, extracting an evolutionary information feature vector and a solvent accessibility feature vector of each amino acid residue; feature fusion, using a linear kernel based multi-core-learning algorithm to carry out evaluation on weight information of the two feature vectors, and according to the weight, carrying out weighted serial combination to obtain a final sample feature vector; using a random downsampling technology to carry out multiple downsampling on non-binding site samples, combining a non-binding site sample subset obtained by downsampling and a binding site sample set to train an SVM, and obtaining a plurality of SVM prediction models; and using a Boosting lifting algorithm to carry out integration on the SVM models, and forming a final prediction model. The method disclosed by the present invention improves model interpretability, effectively reduces the size of the training set, and improves the prediction precision of the model.
Owner:NANJING UNIV OF SCI & TECH
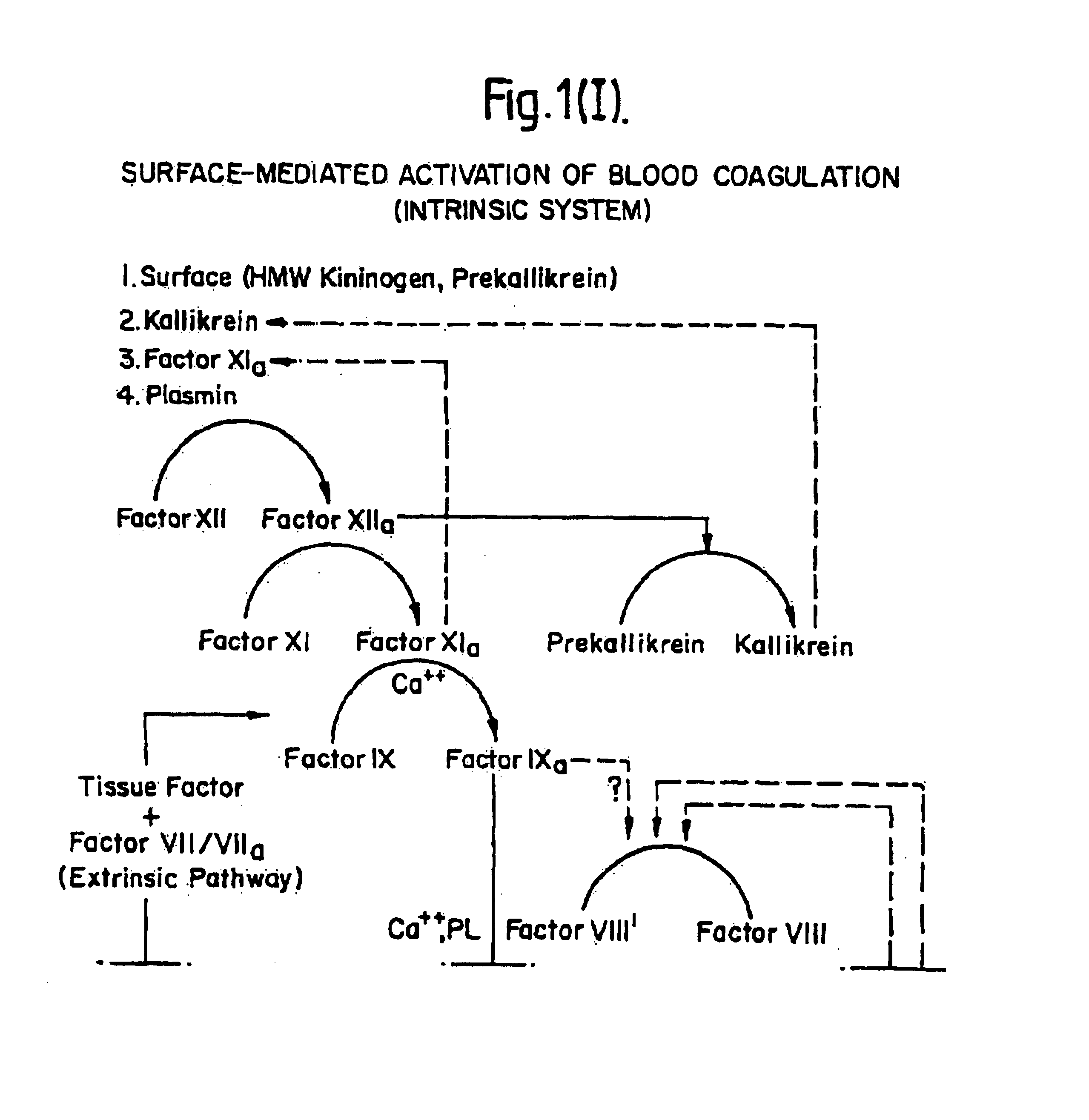
Methods and Deoxyribonucleic acid for the preparation of tissue factor protein
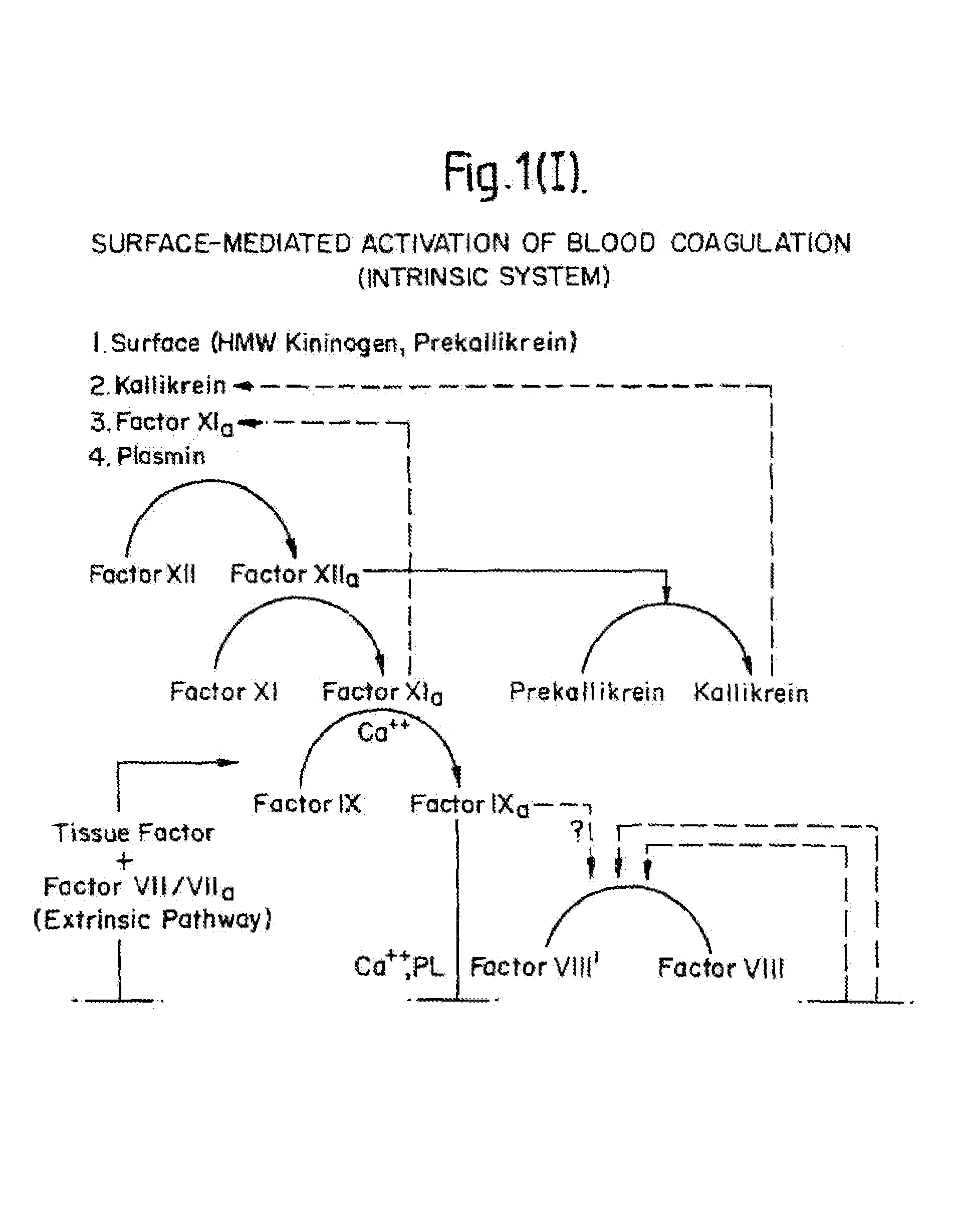
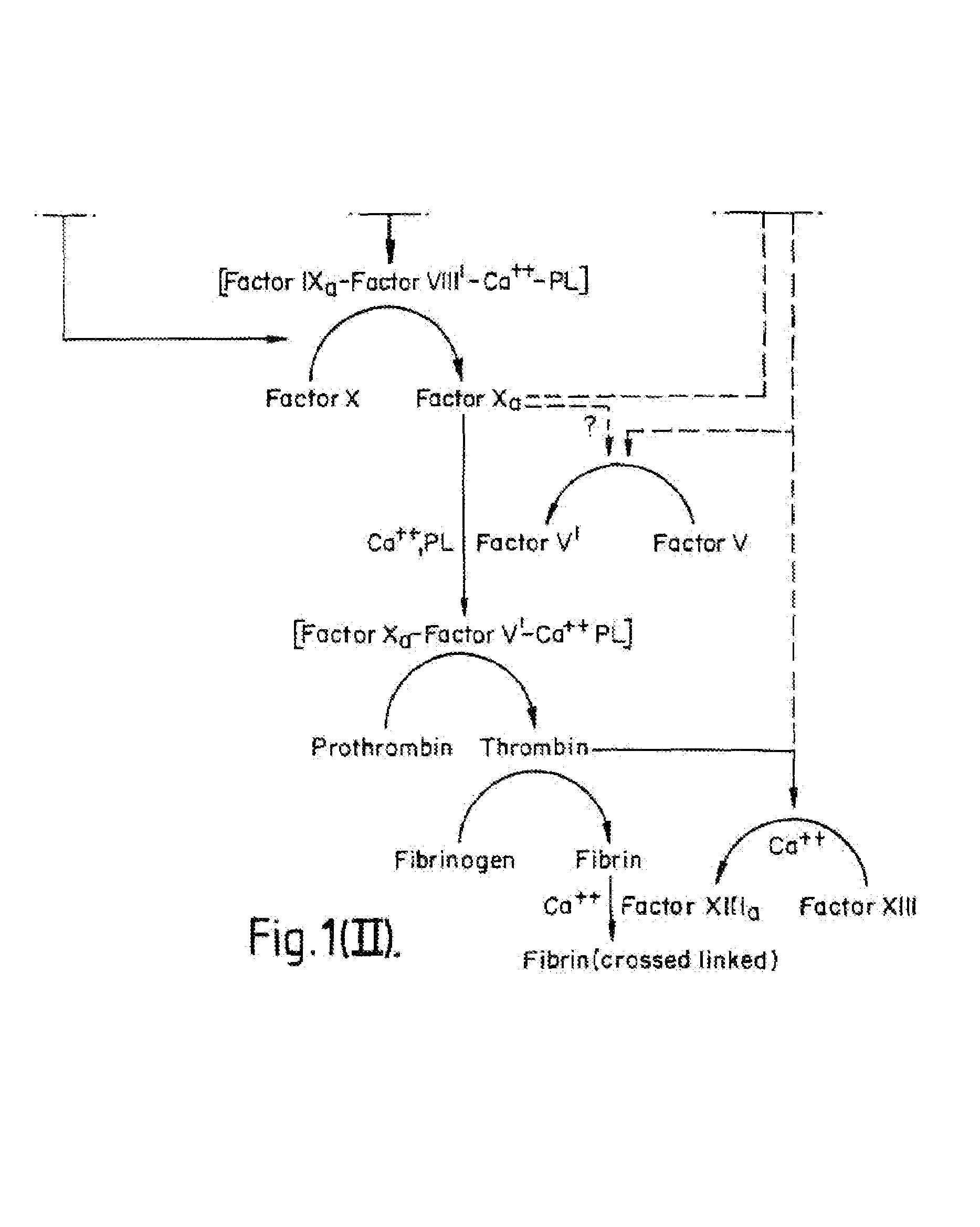
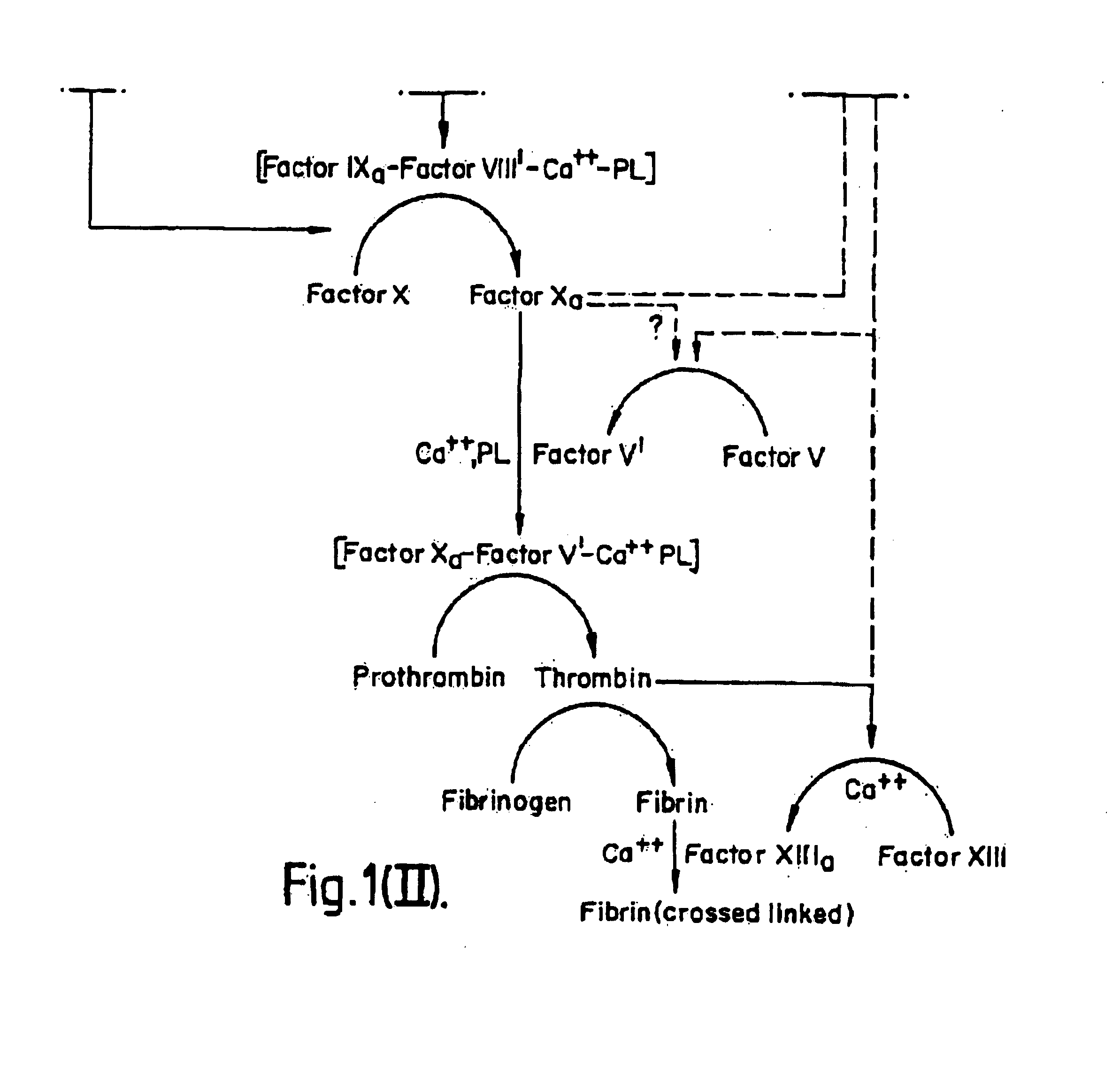
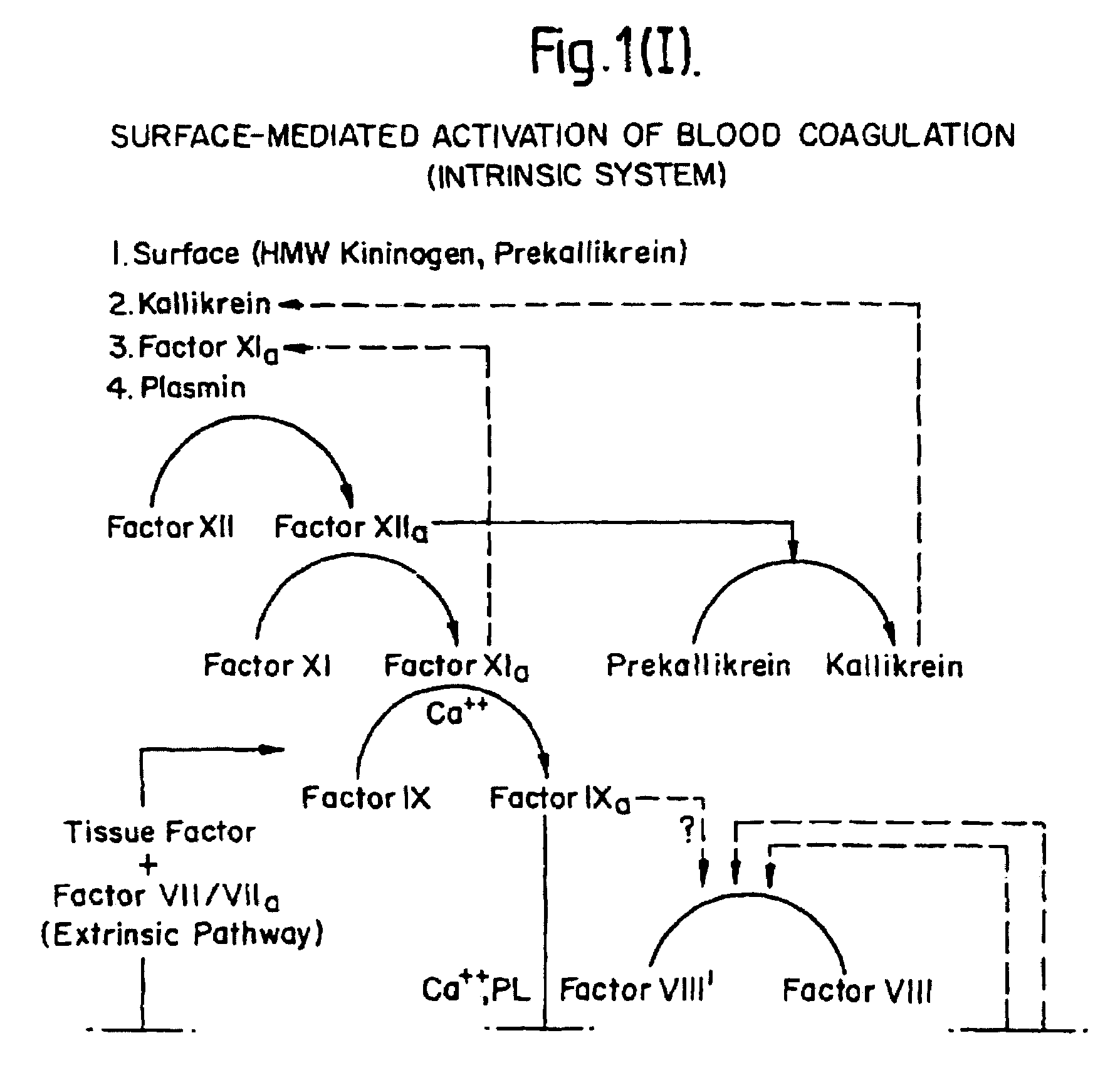
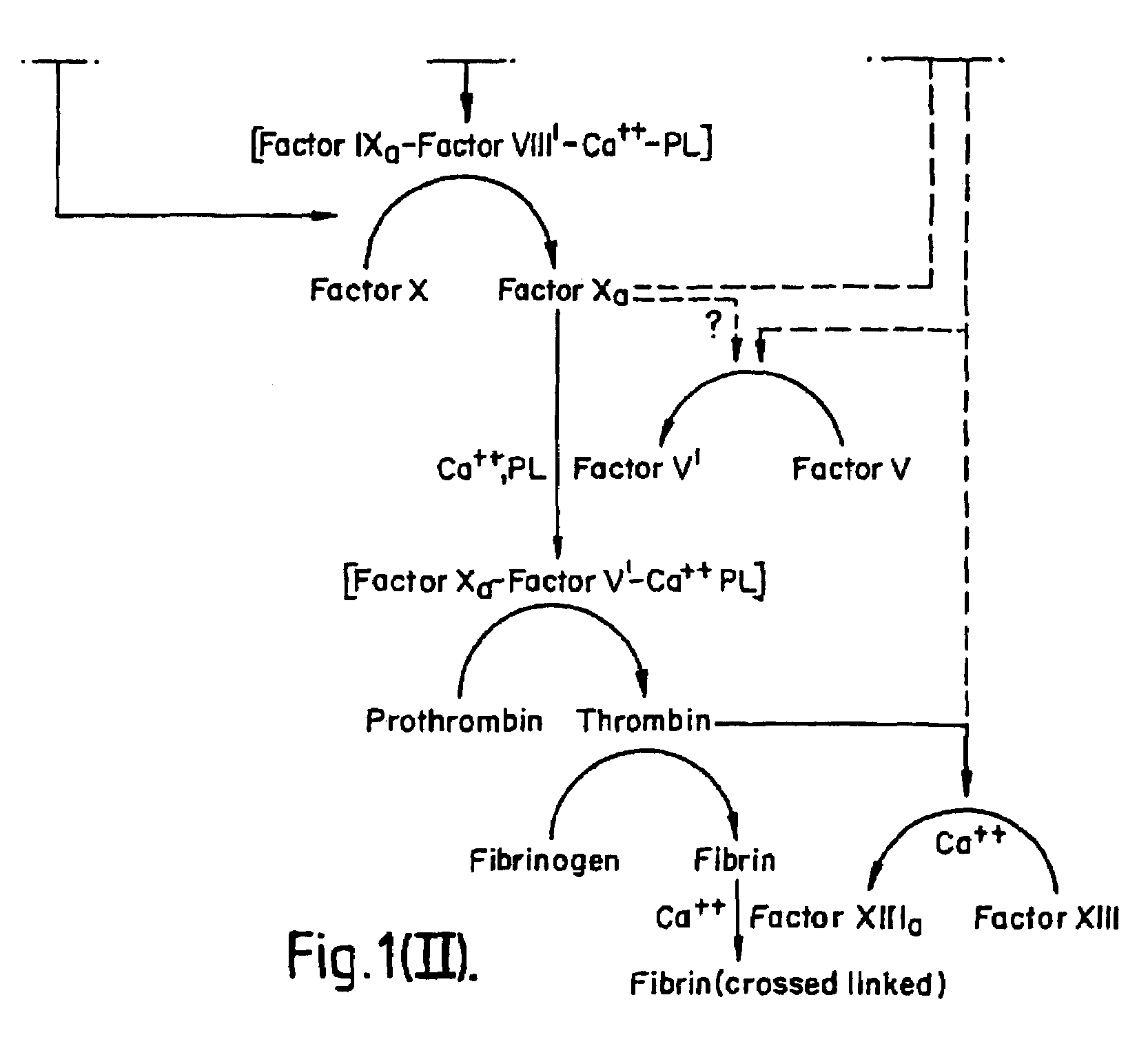
InactiveUS7084251B1Difficult to identifyIsolation of difficultDepsipeptidesPeptide preparation methodsTissue factorCoagulation Disorder
DNA isolates coding for tissue factor protein and methods of obtaining such DNA and producing tissue factor protein using recombinant expression systems for use in therapeutic composition for the treatment of coagulation disorders.
Owner:GENENTECH INC
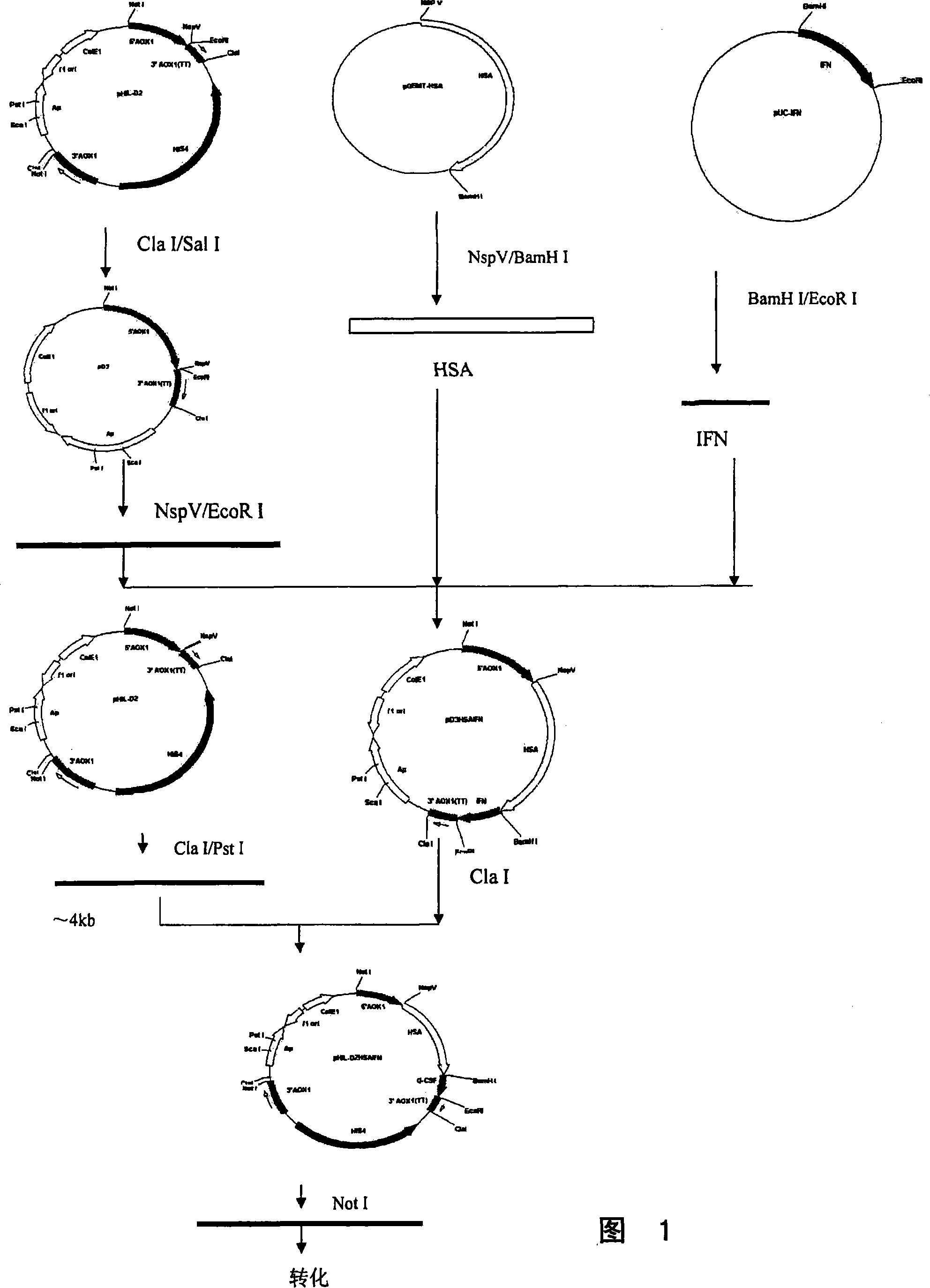
Fusion protein for seralbumin and interferon
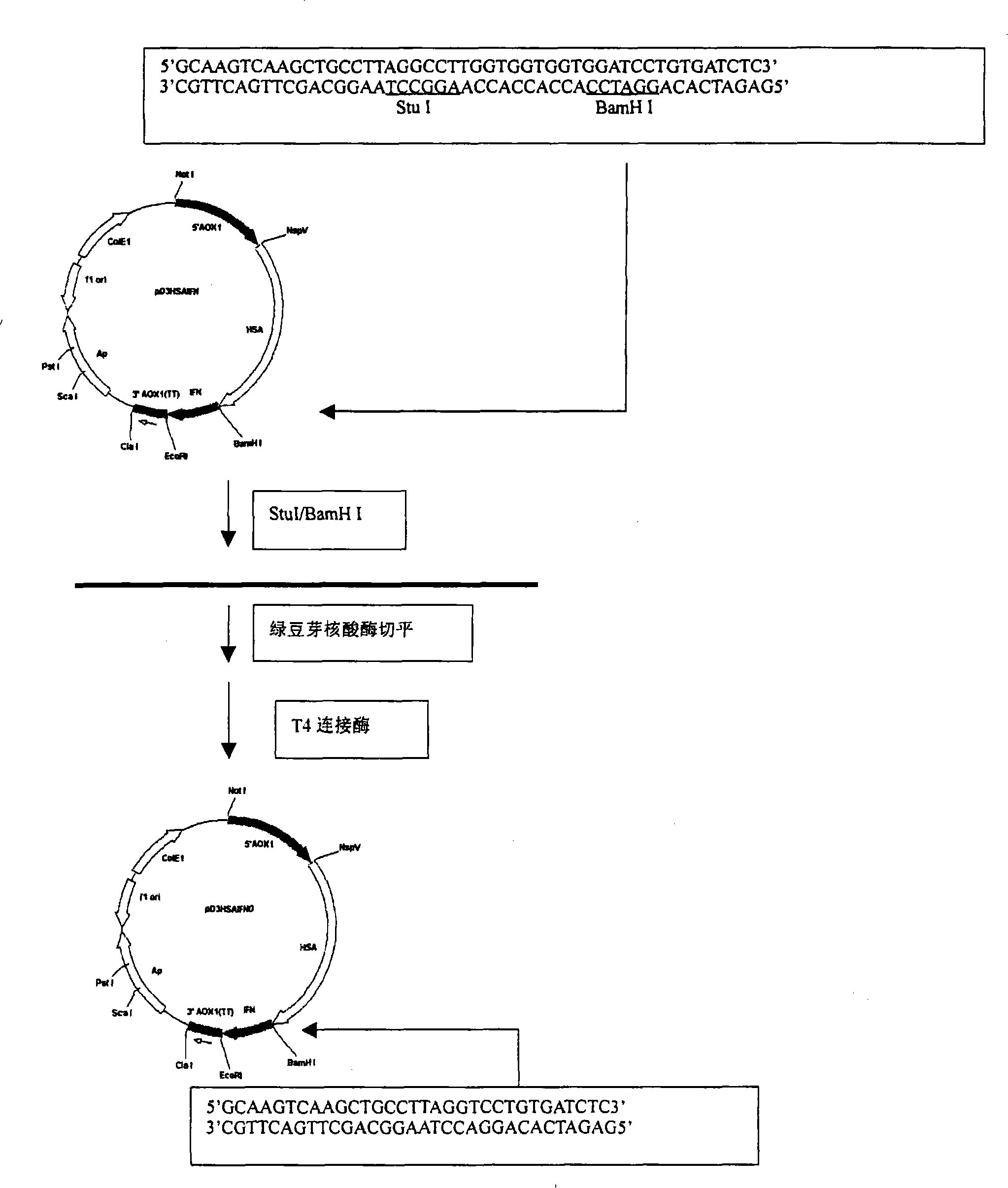
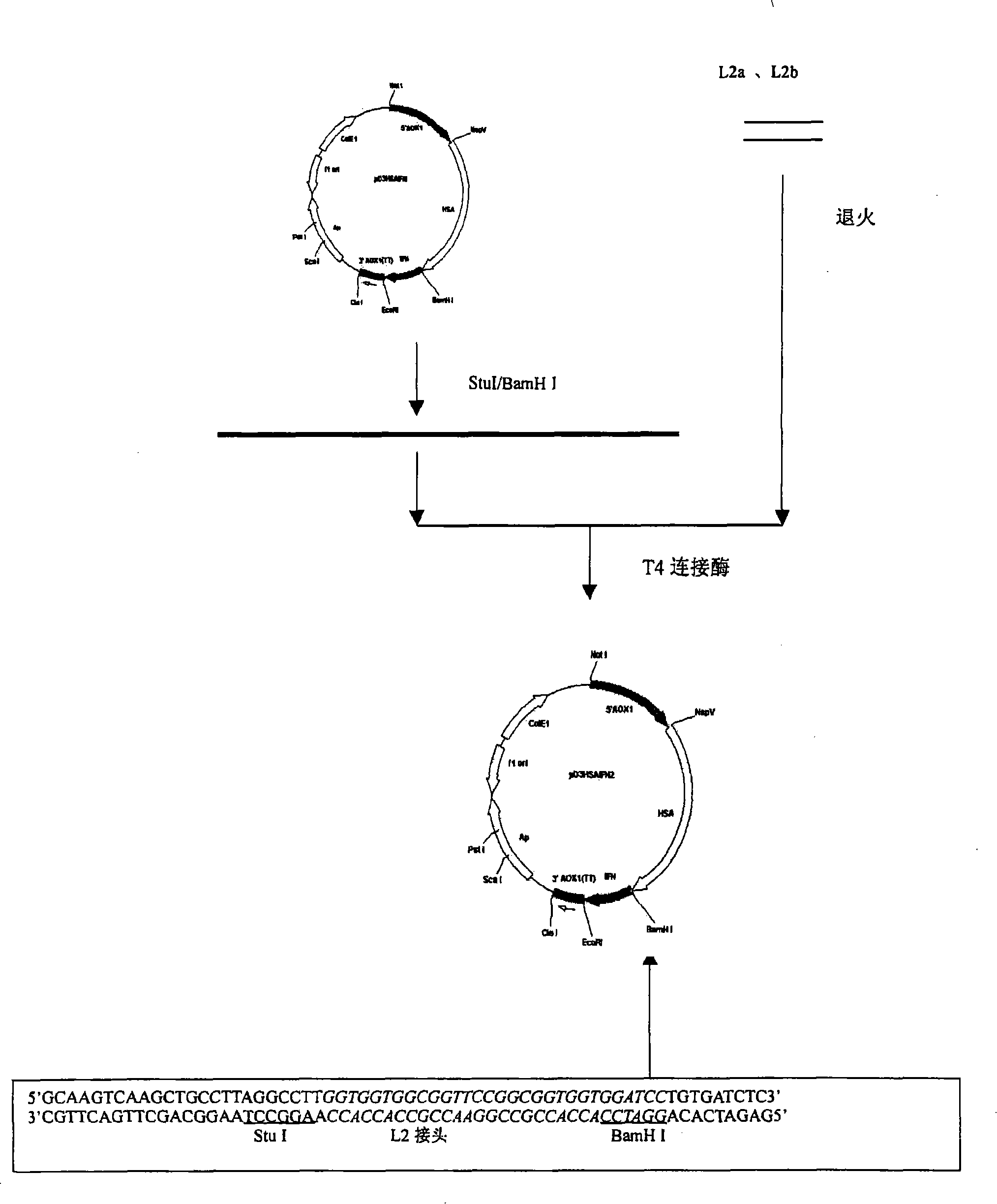
InactiveCN101200503ARetain activityProlonged plasma half-lifeFungiAlbumin peptidesHalf-lifePlant cell
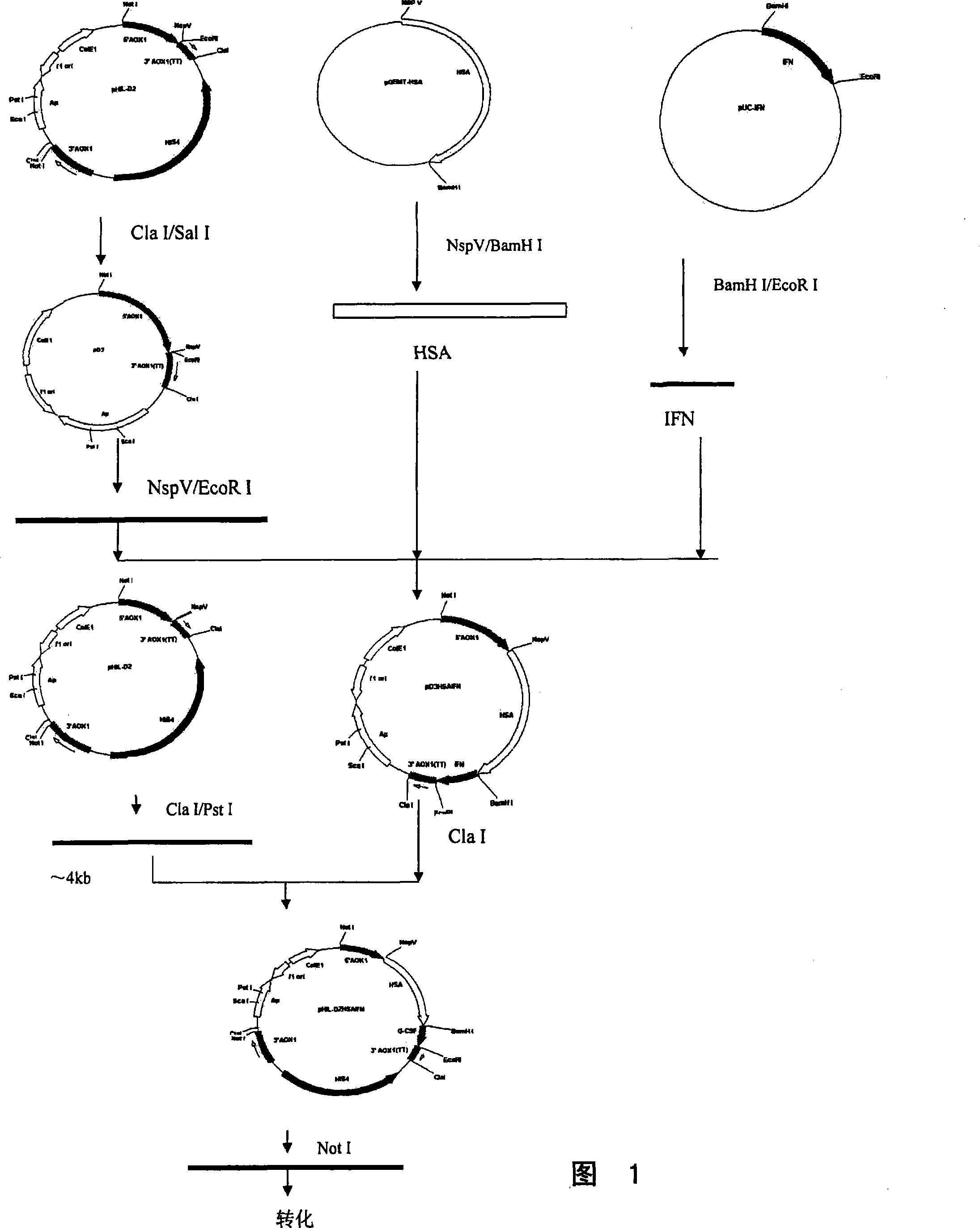
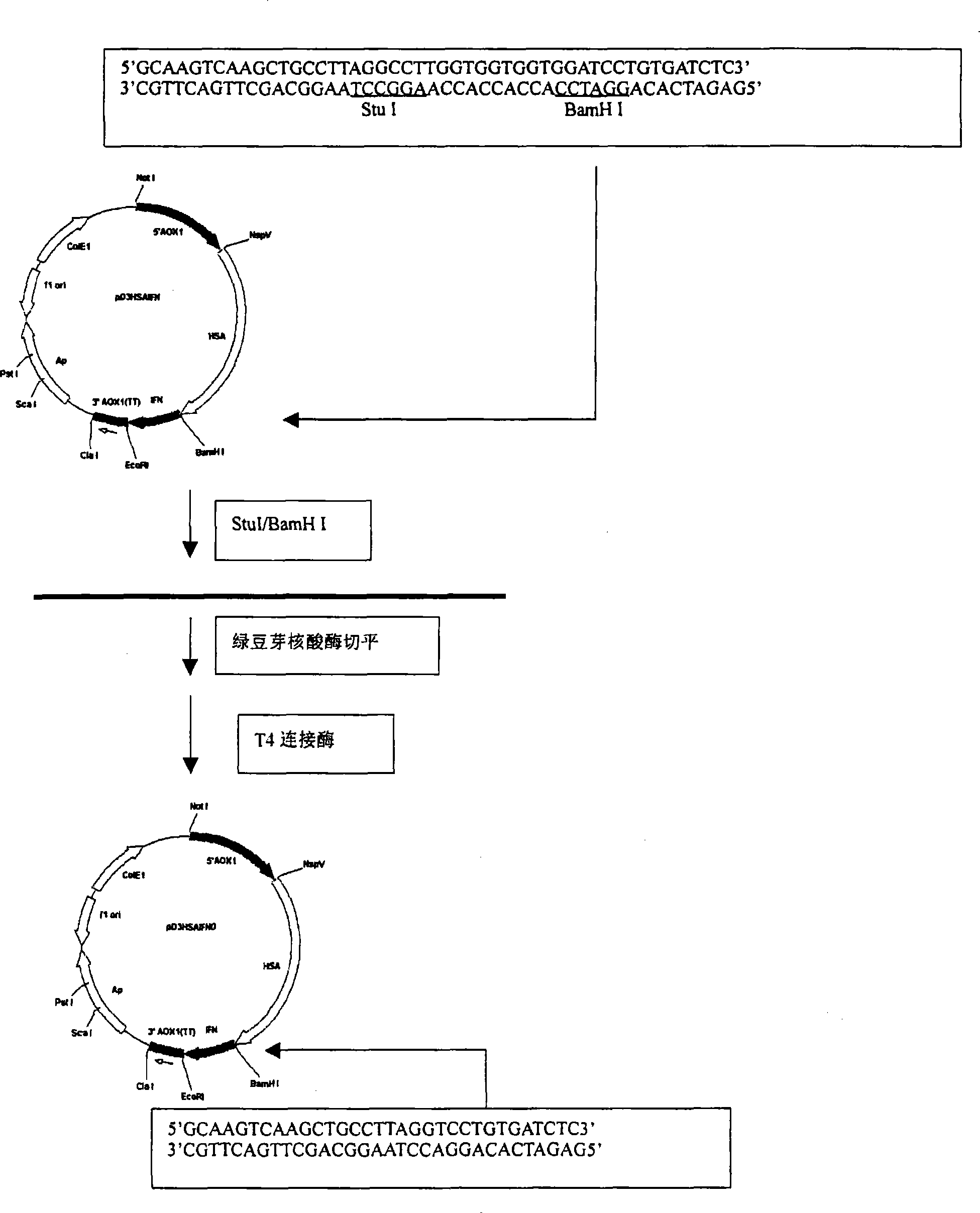
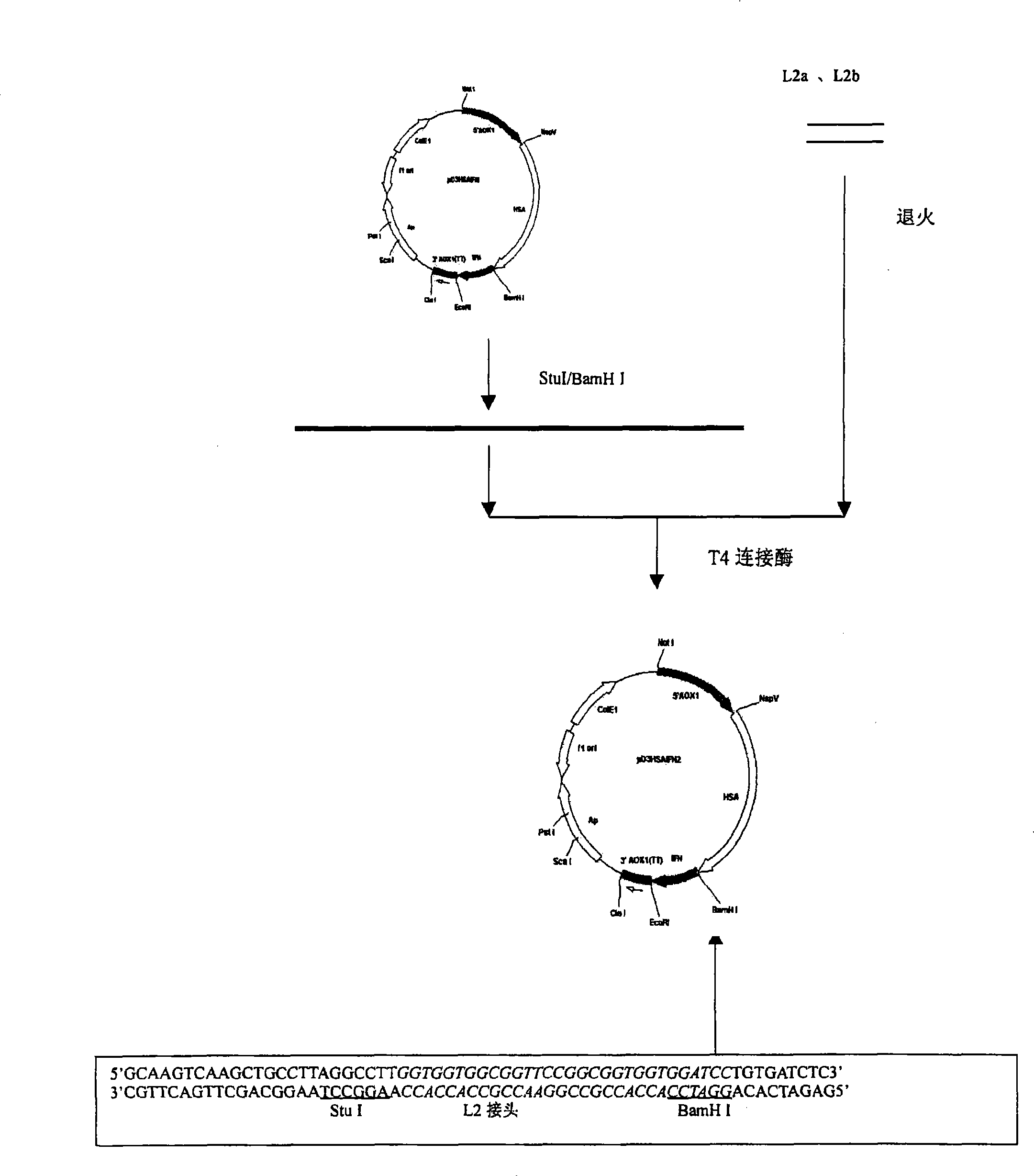
The present invention discloses a fusional protein of a serum albumin and an interferon, the coding of the fusional protein DNA sequence, as well as bacteria, barm, animal cell and plant cell of the DNA sequence. The fusional protein provided by the present invention includes a first district with at least 85 percents congeneric sequence of the artificial serum albumin and a second district with at least 85 percents congeneric sequence of the artificial interferon. Without changing the property of the fusional protein, the fusional protein can implement the replacement, deficit or addition of exceptional amide acid residue. A connecting peptide is arranged between the first district and the second district of the fusional protein provided by the present invention; the general formula of the connecting peptide is [GlyGly GlyGlySer]n and n is an integer from 1 to 10. The fusional protein provided by the present invention with the property of artificial interferon and prolonged half-value period will be widely applied in the medicine field.
Owner:INST OF BIOENG ACAD OF MILITARY MEDICAL SCI OF THE CHINESE +2
Firefly luciferase and encoding gene and obtaining method thereof
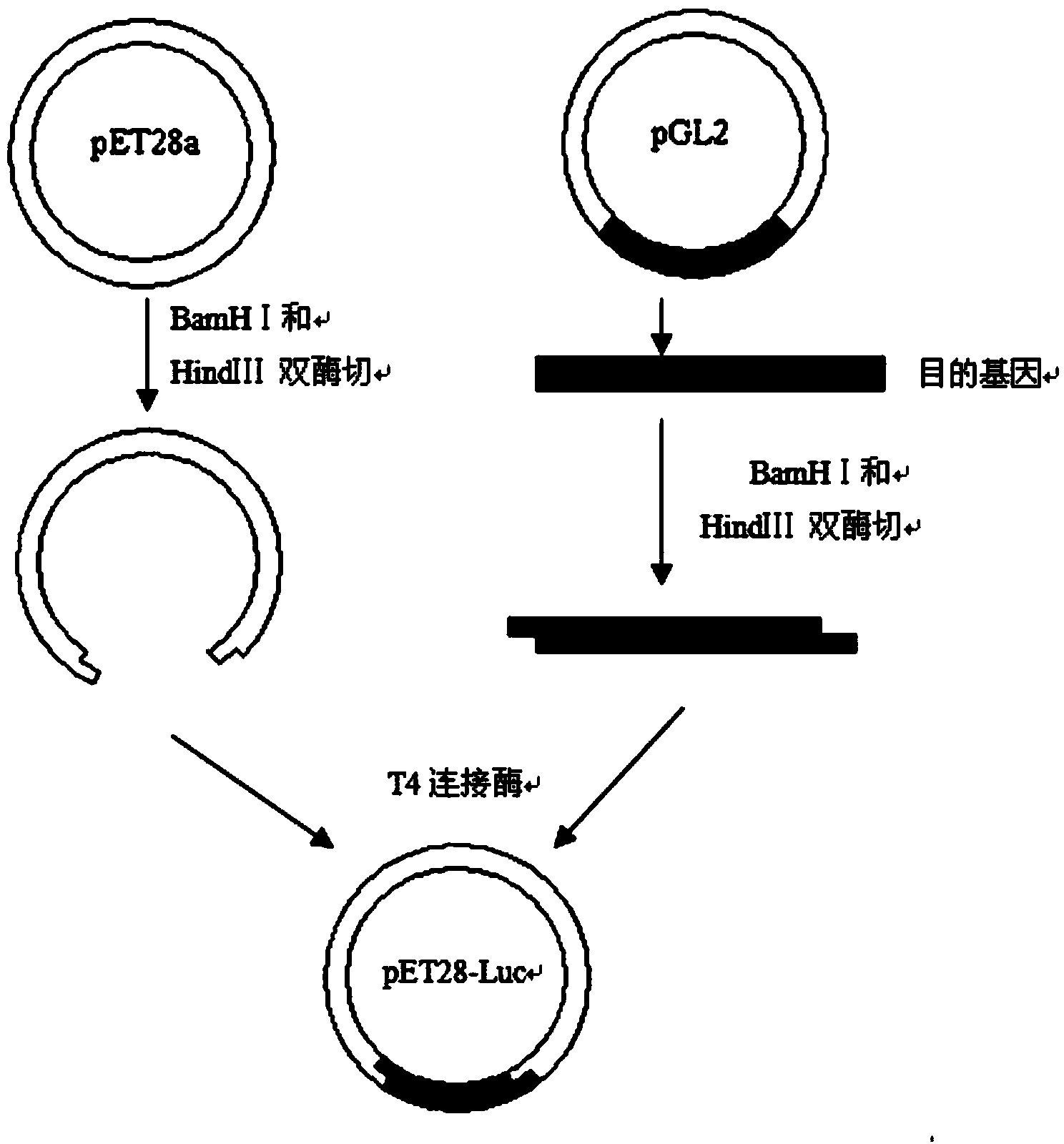
ActiveCN103409380AHigh catalytic activityHigh activityBacteriaMicroorganism based processesHeat stabilityWild type
The invention relates to firefly luciferase and an encoding gene and an obtaining method thereof. The defect that the original firefly luciferase in North America is poor in heat stability and cannot be used at a high temperature is overcome; and the original firefly luciferase in North America is mutated, so that mutative enzyme of which the heat stability is obviously improved is obtained; and the application value is improved. An amino acid sequence is SEQ ID NO.1; and a protein deoxyribonucleic acid (DNA) sequence in encoding is SEQ ID NO.2. An encoding gene and a preparation method of firefly luciferase mutant in North America are provided. By the method, a recombinant expression vector containing a wild firefly luciferase gene is built; and the protein is purified by using an affinity chromatography method. The method is simple and feasible, simple to operate, and low in cost.
Owner:TIANJIN UNIV
Efficient transgenosis method with mediation of transcription activator-like effector protein
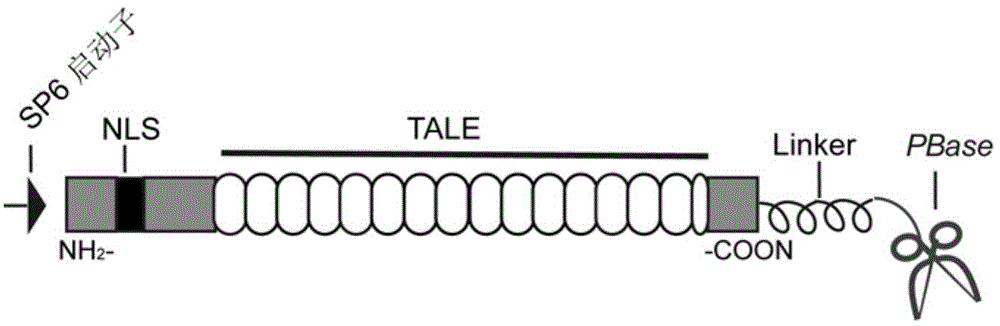
ActiveCN104651409ASimplify the process of experimentationImprove transgenic efficiencyMicroinjection basedFermentationTransgeneMicroinjection
The invention discloses an efficient transgenosis method with mediation of a transcription activator-like effector (TALE) protein. The method comprises the steps that a helper plasmid containing a transcription activator-like effector protein DNA (Deoxyribonucleic Acid) sequence and piggyBac transposon (PBase) DNA sequence is constructed by adopting a molecular biological technique, and transcribed to mRNA (Messenger Ribonucleic Acid) in vitro; mRNA of the helper plasmid and DNA of a piggyBac transposon plasmid containing an exogenous gene are imported to a fertilized egg of an animal by adopting a microinjection method; a transgenic animal of which the exogenous gene can be stably inherited and expresses is obtained; and a transgenosis positive individual is screened out according to a marker gene or the expression characteristic of the gene. With the adoption of the method, a piggyBac transposon can be efficiently inserted into a genome of a host organism by the action of a TALE-PBase fusion protein; the transgenosis efficiency mediated by the piggyBac transposon can be improved remarkably; and helps are offered to researches such as transgenic animal acquirement and insertion mutagenesis.
Owner:ZHEJIANG UNIV
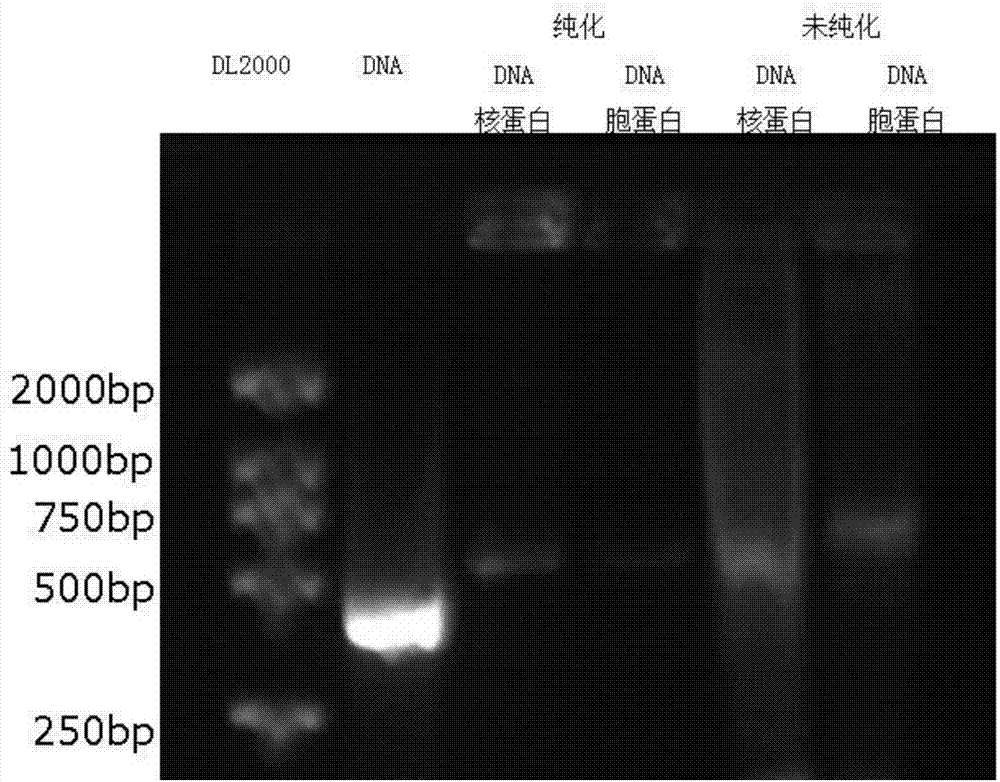
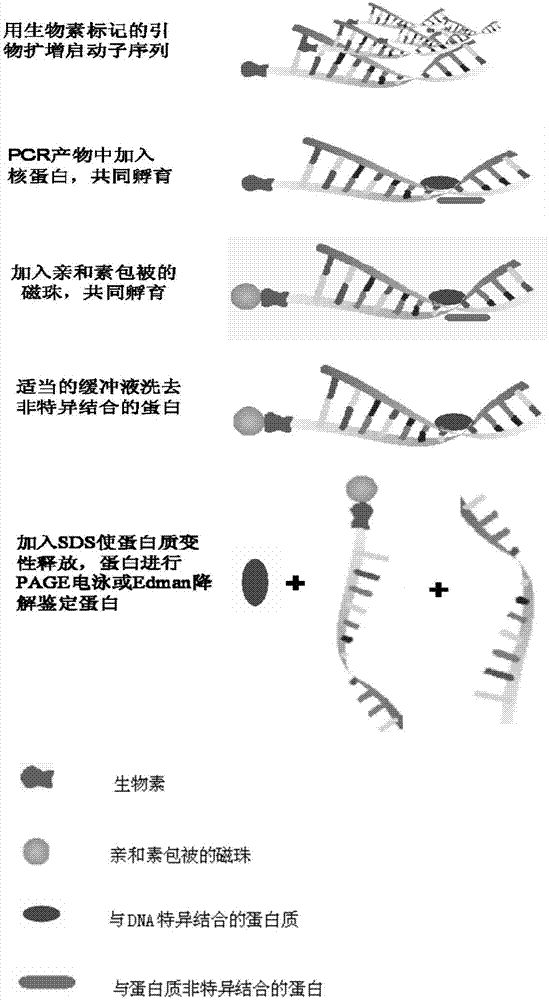
Method for separating DNA (deoxyribonucleic acid) binding protein and accurately positioning DNA binding site
InactiveCN104725465AAccurately locate the site of actionEasy to introduceMicrobiological testing/measurementPeptide preparation methodsMagnetic beadBinding site
The invention provides a method for separating DNA (deoxyribonucleic acid) binding protein and accurately positioning DNA binding site, which comprises the following steps: DNA binding protein separation: carrying out PCR (polymerase chain reaction) amplification on a DNA segment to be researched by using biotin-labeled primers, sequentially adding nucleoprotein or cytoplast protein and avidin-coated magnetic beads, co-incubating, washing off nonspecific binding protein, and adding SDS (sodium dodecylsulfate) to denature and release the protein; and protein DNA binding site positioning: by using the DNA segment to be researched as a template, carrying out PCR reaction by using different primers to obtain overlapping DNA segments, and positioning the DNA binding site of the isolated protein according to the affinity difference of the overlapping DNA segments for the isolated protein.
Owner:CAPITAL UNIVERSITY OF MEDICAL SCIENCES
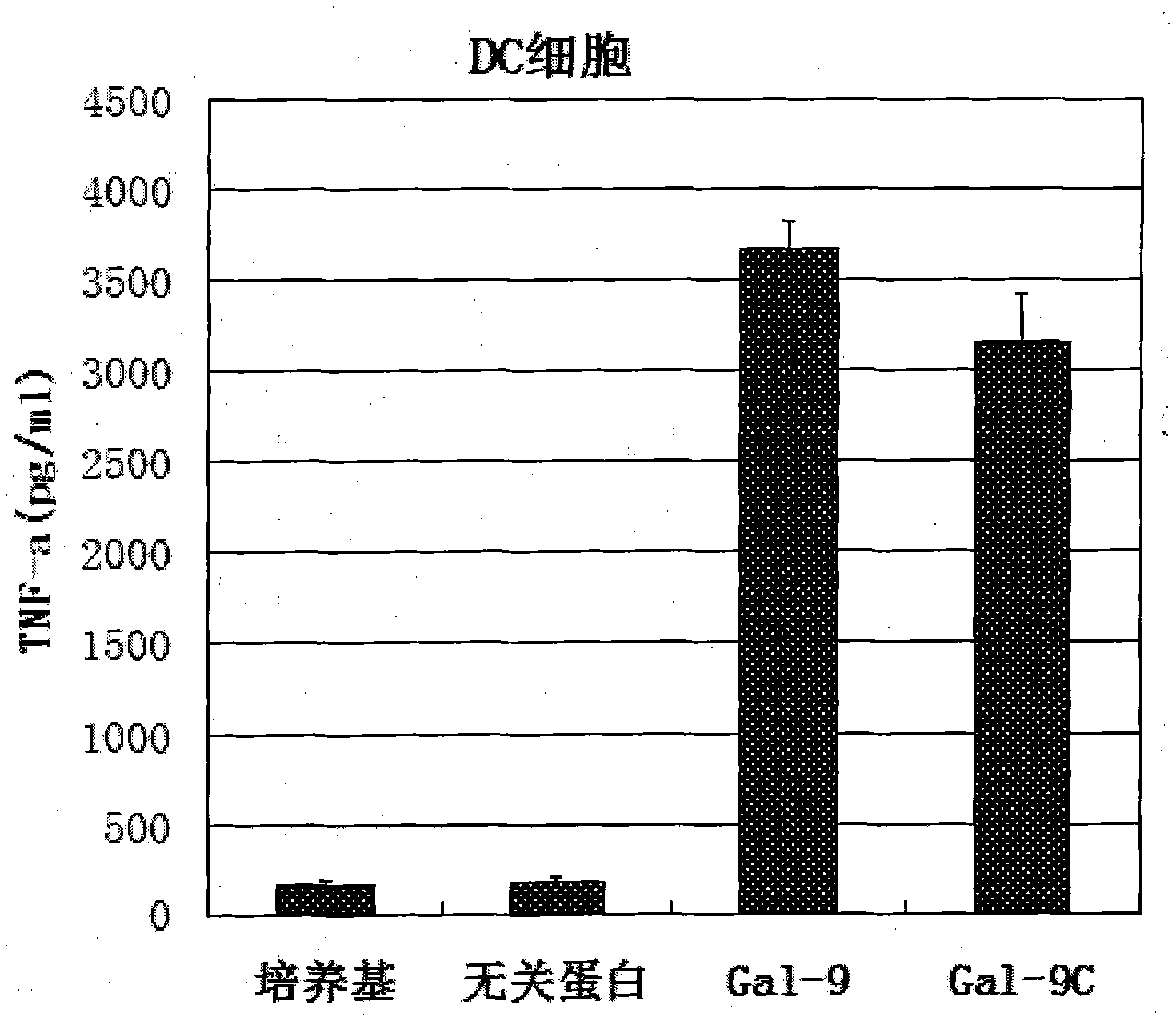
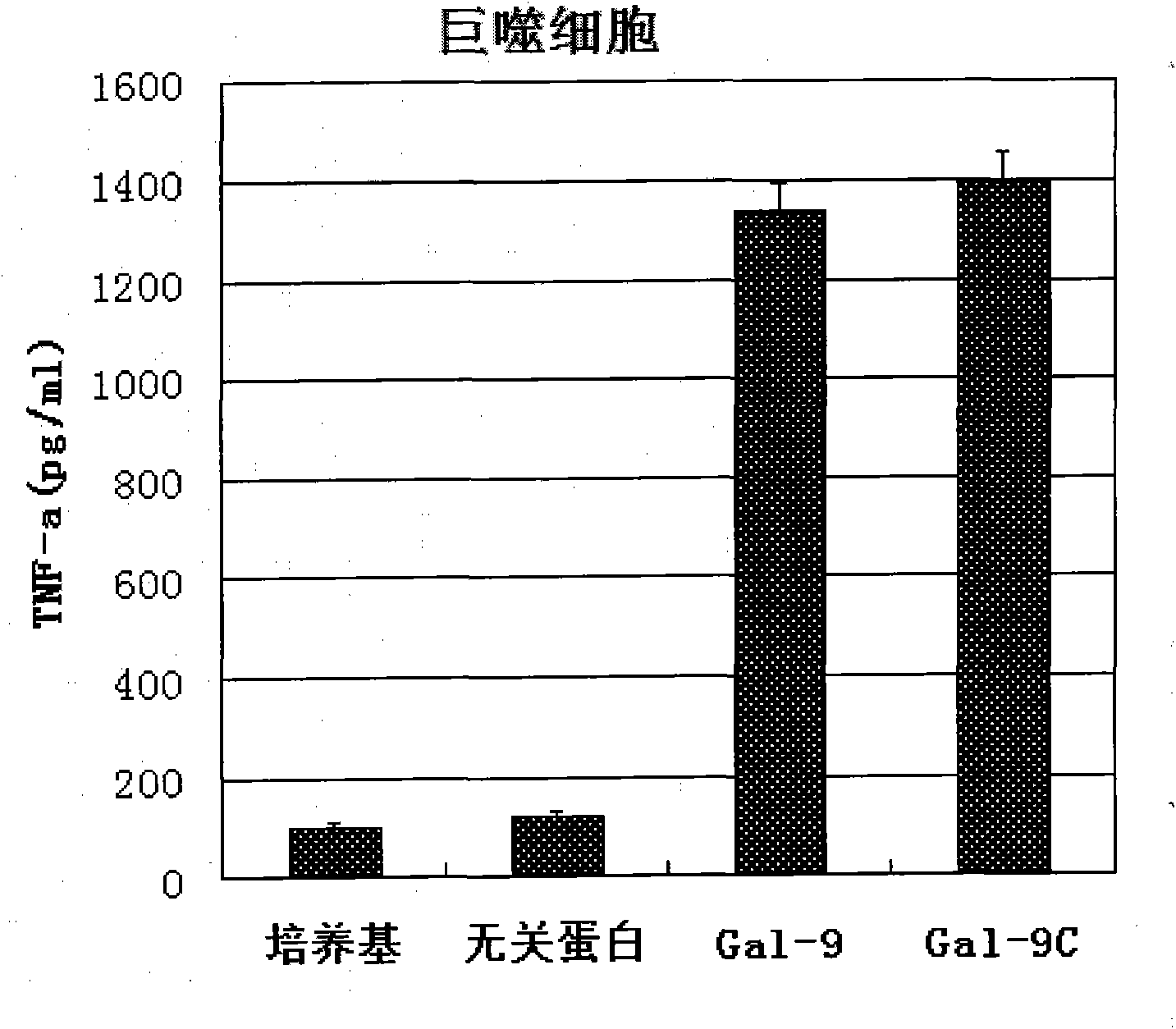
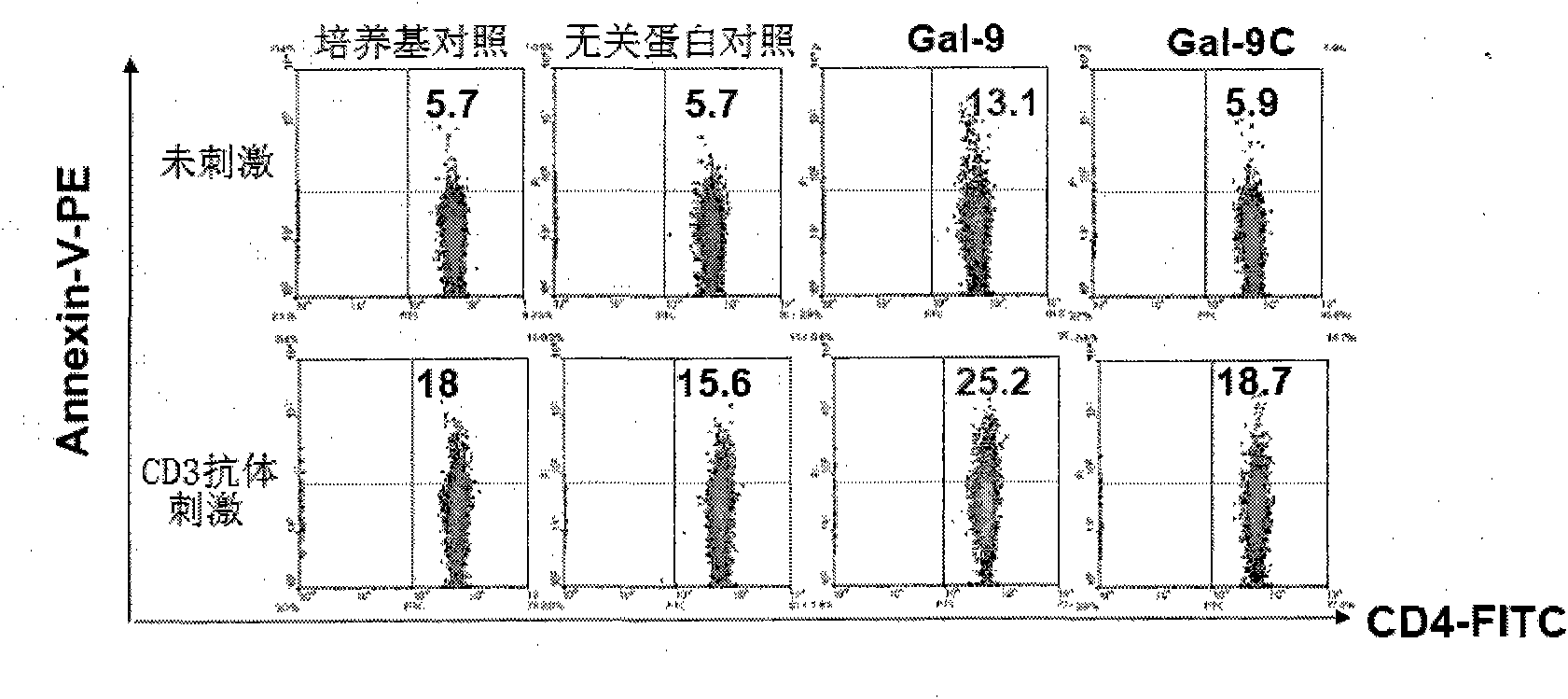
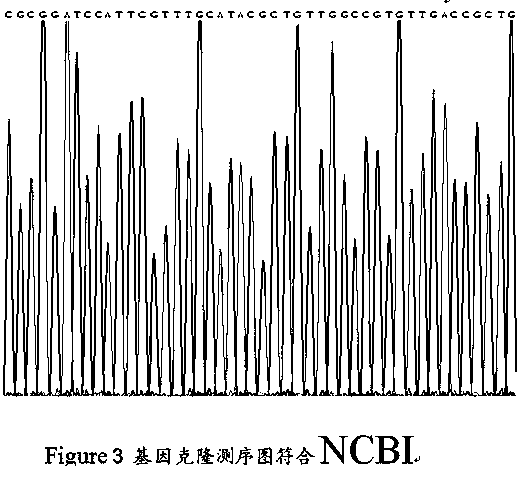
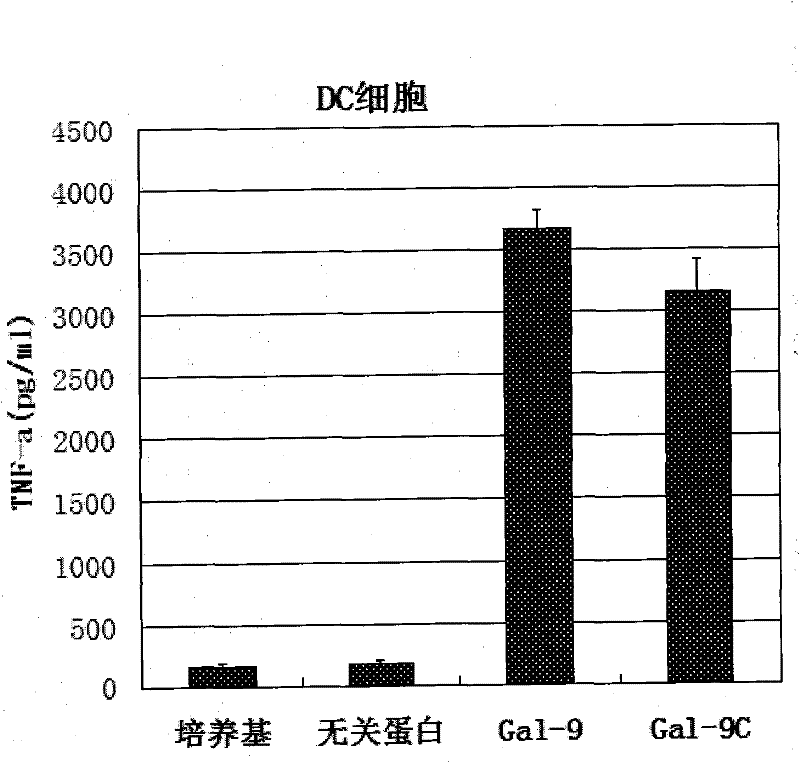
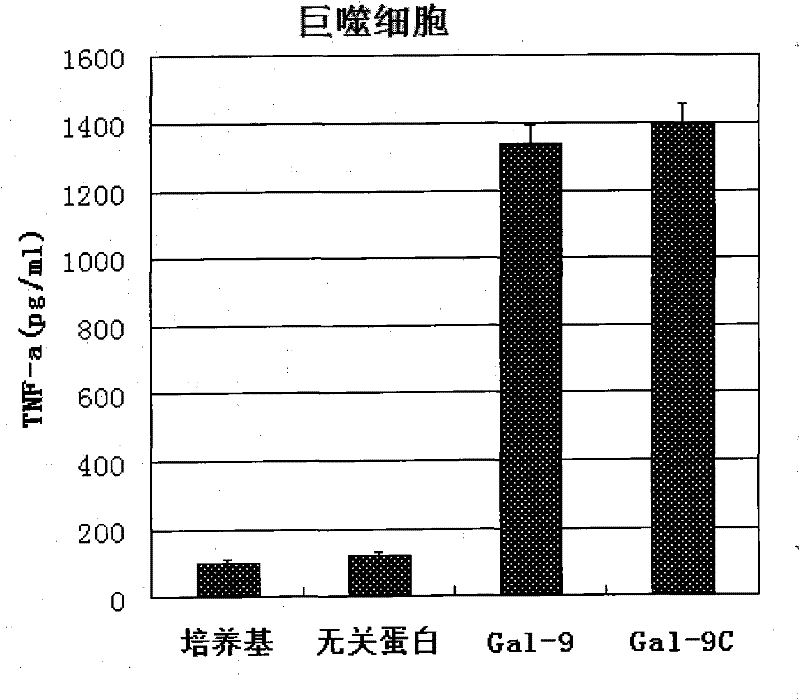
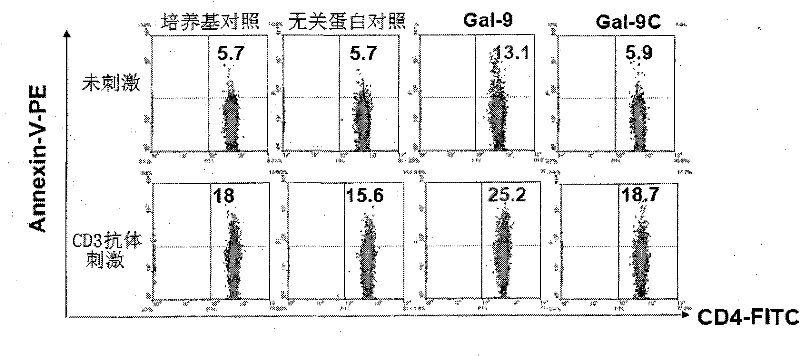
Preparation and application of human galectin-9 deletant for enhancement of immune response
InactiveCN101792489AImprove the activation effectImprove immunityFungiBacteriaImmunosuppressive effectBiological activation
The invention discloses a preparation method of human galectin-9 deletant for enhancement of immune response, which adopts an expression vector with a protein DNA sequence and a host cell transformed with the expression vector. The invention also provides application of the human galectin-9 deletant in improvement of cellular immunity. The protein has the function of promoting the activation of DC cells and macrophages, the demonstration of the function of the protein is the enhancement of the cellular immune response, and the immunity is suppressed in case of lack of the human galectin-9 holoprotein.
Owner:INST OF BASIC MEDICAL SCI ACAD OF MILITARY MEDICAL SCI OF PLA
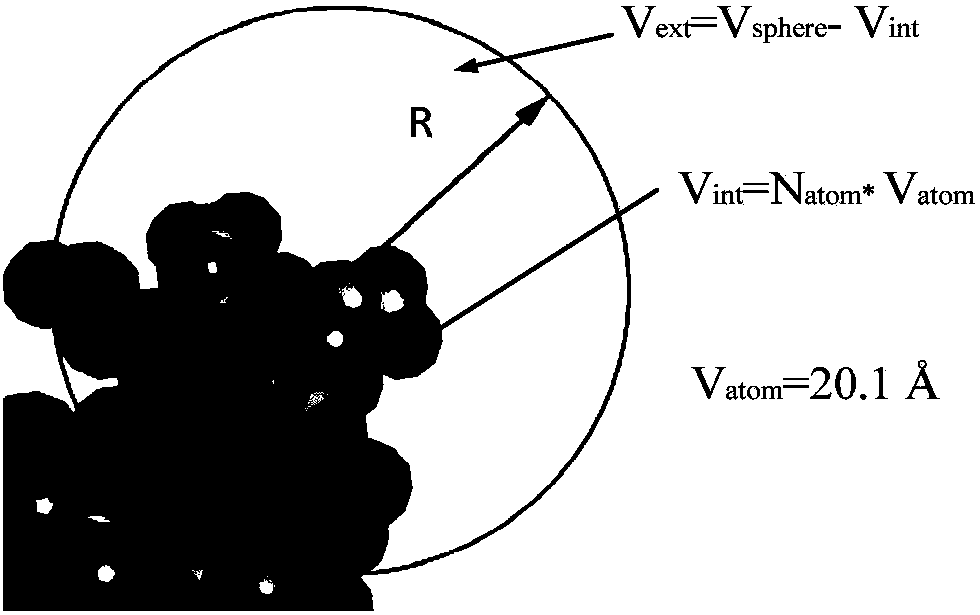
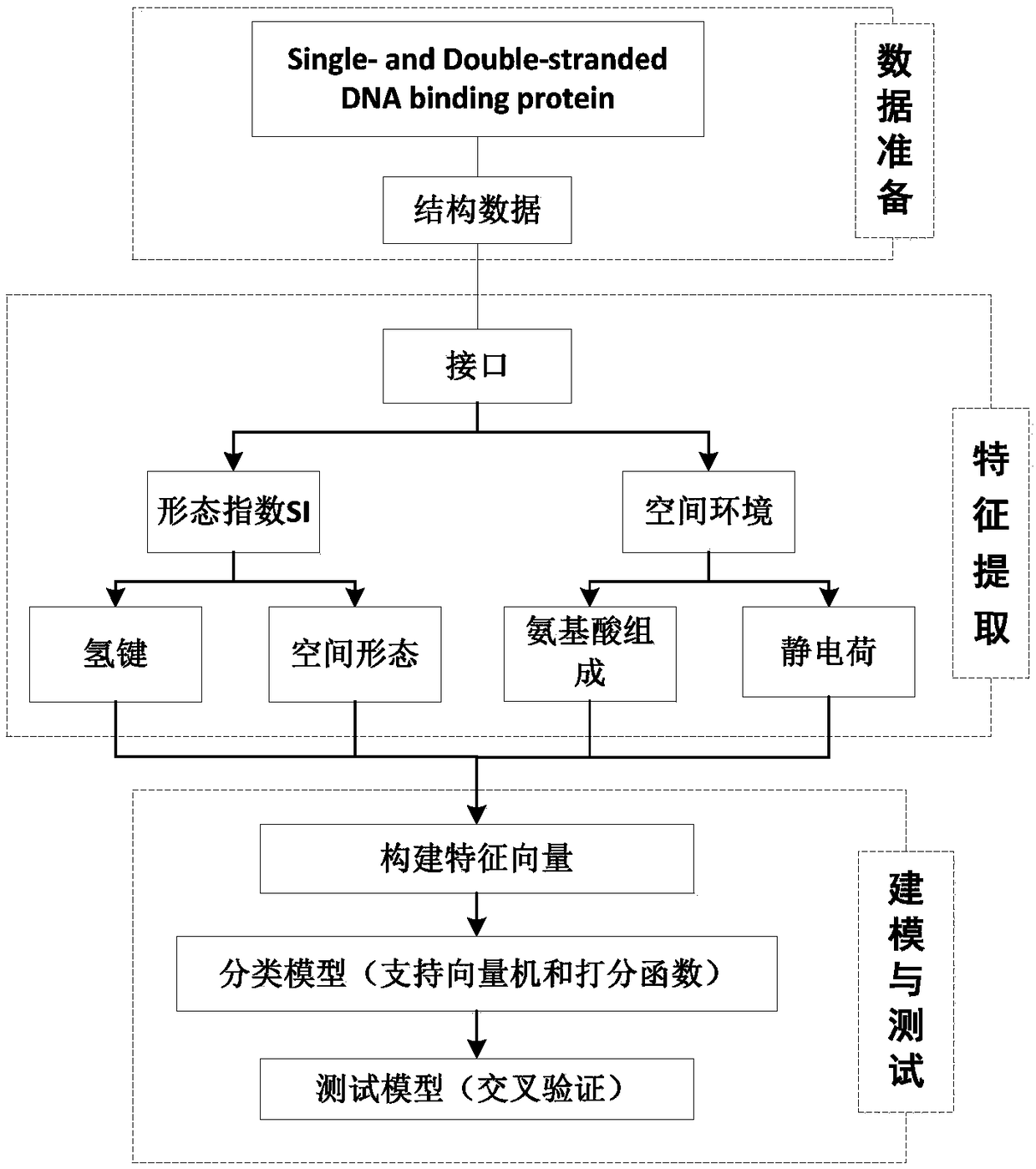
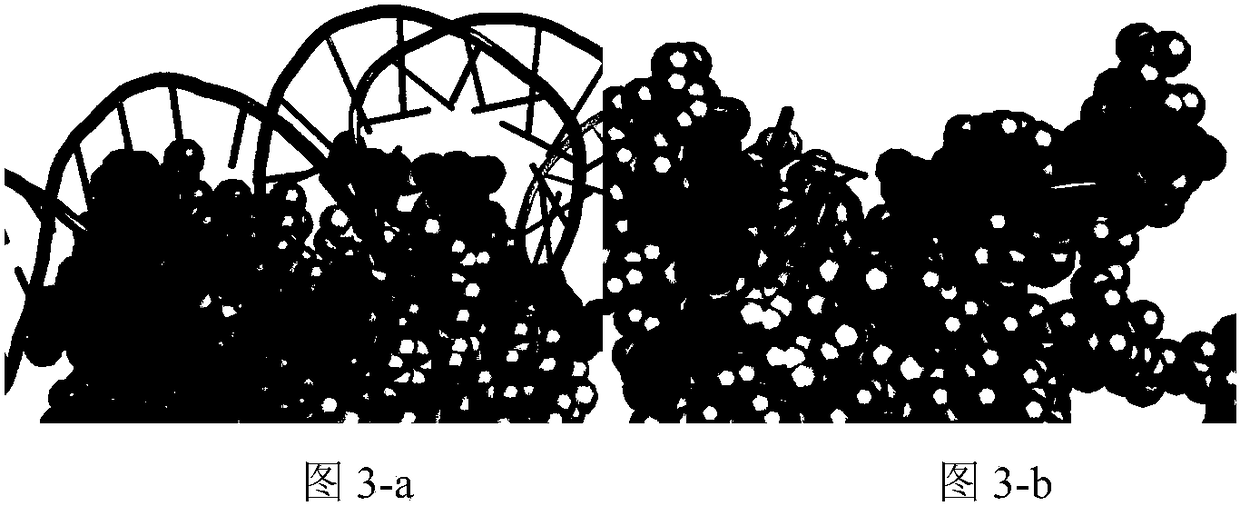
A method for analyzing the geometric structure characteristics of DNA bin protein interface
InactiveCN109101784AIn-depth understanding of the binding mechanismSpecial data processing applicationsBinding siteSingle strand dna
The invention relates to an analysis method for the geometrical structure characteristics of DNA binding protein interface, belonging to the technical field of biological computing. The present invention first derives proteins-DNA binding site information from proteins-DNA complex dataset; and then based on the protein-DNA Binding Site Information, residue Morphological Index and Spatial Environment of proteins-DNA Binding Sites are determined; finally, the morphological index and spatial environment of the identified protein-DNA binding sites are used as feature vectors to classify the obtained feature vectors by using classification model. As the binding specificity of double-stranded DNA binding protein (DSBs) and single-stranded DNA binding proteins (SSBs) is analyzed from the angles of the morphological structure of the residues and the characteristics of the space environment, the invention is conducive to the in-depth understanding of the proteins, and the binding specificity ofthe double-stranded DNA binding proteins and the single-stranded DNA binding proteins is analyzed. Based on the DNA binding mechanism, the present invention can also be applied to proteins-RNA, protein-Protein and other fields.
Owner:HENAN NORMAL UNIV
Method for determining transcriptional control complex
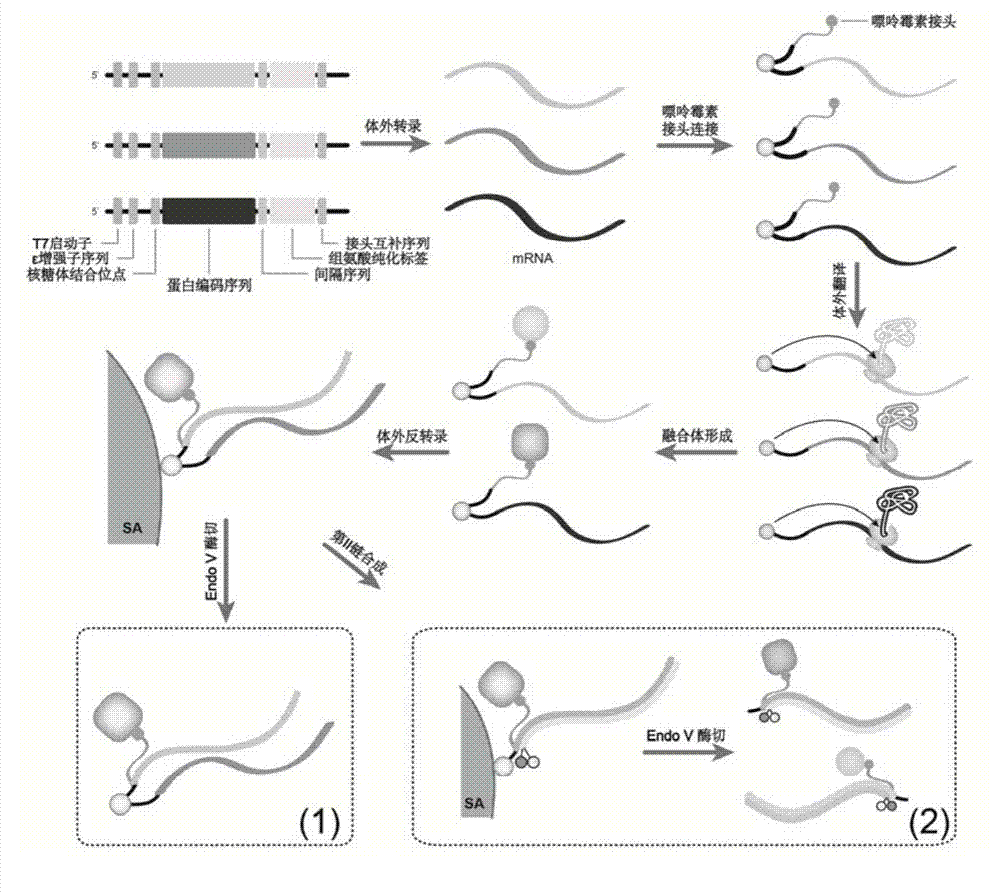
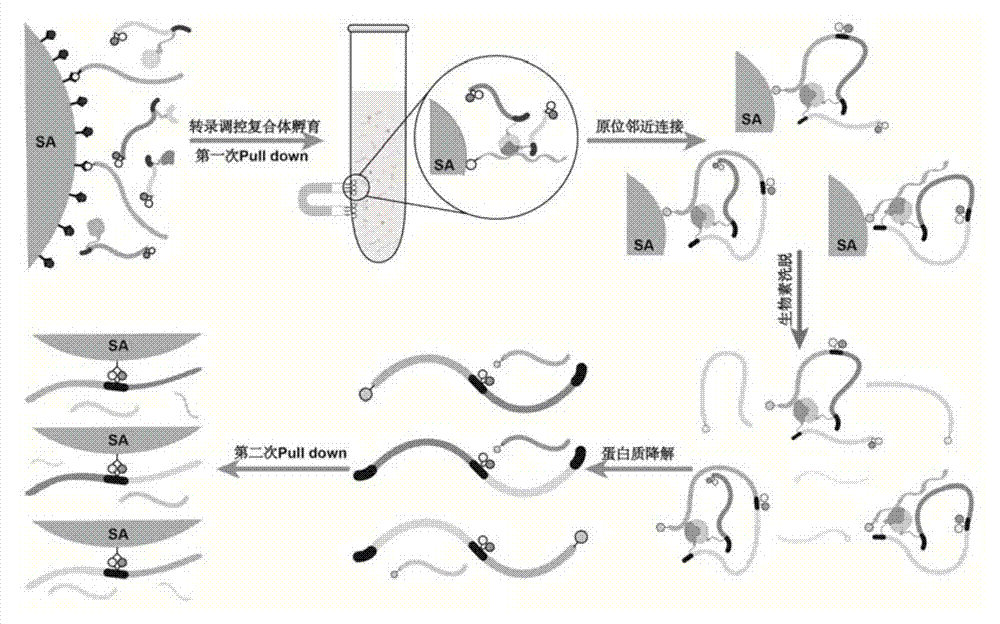
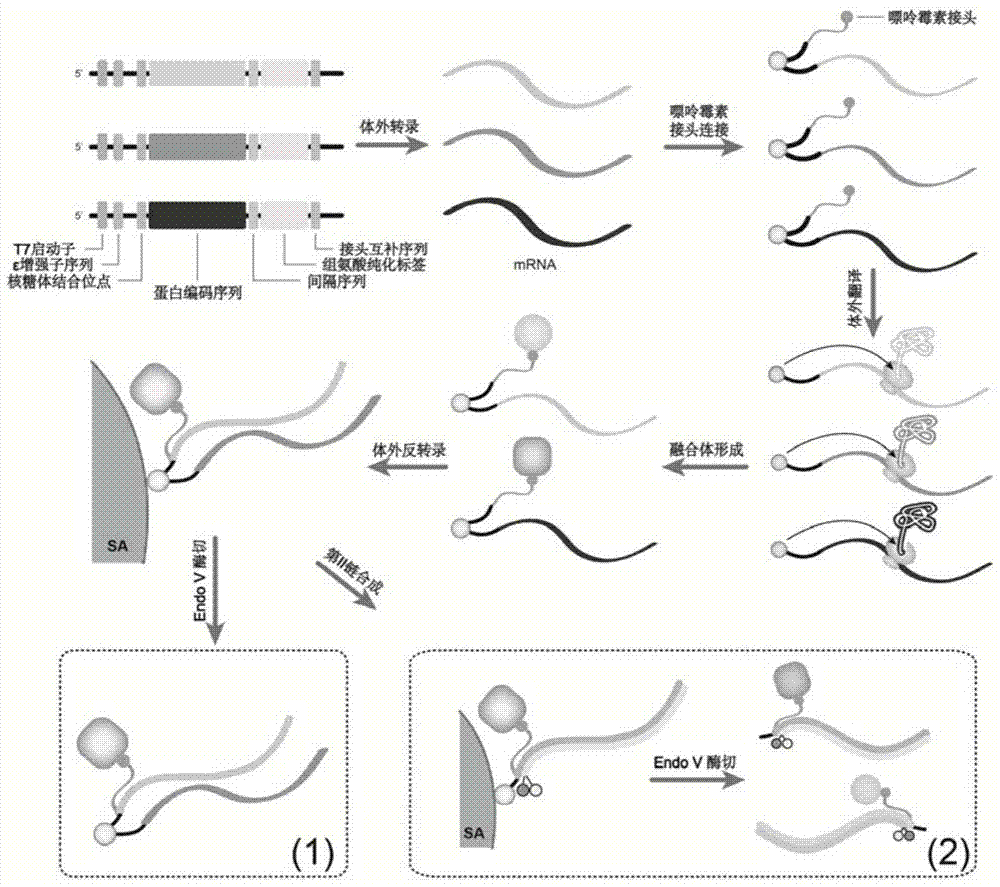
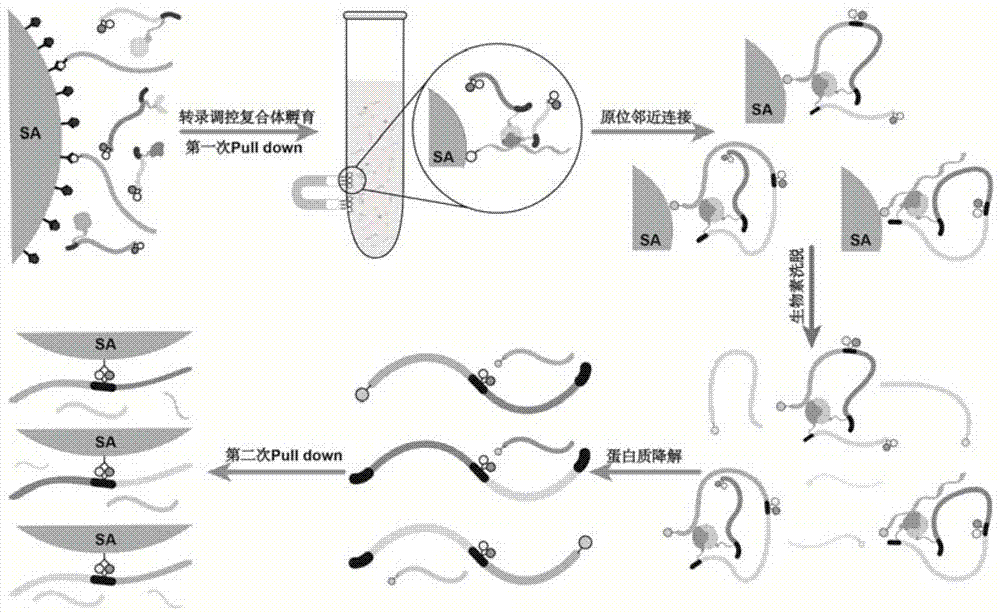
ActiveCN104774923AIncrease library capacityMitigate incompatibilitiesMicrobiological testing/measurementDna interactionProtein–DNA interaction
The invention relates to a method for determining a transcriptional control complex, and belongs to the field of molecular biology. The method for determining the transcriptional control complex includes the following steps: preparation of a bait DNA sequence, preparation of a DNA sequence for encoding a bar code protein, construction of a bar code protein library, screening of the transcriptional control complex, and in situ adjacent connection capture of the transcriptional control complex, and the identification of the transcriptional control complex. The invention combines cDNA display technology and the adjacent connection technology to establish a novel method for determining the transcriptional control complex; the determination of transcriptional control complex involves simultaneous determination of a variety of interaction effects of protein-protein and protein-DNA; and the method can realize simultaneous determination of a variety of interaction effects of protein-protein and protein-DNA in a same experimental system, and effectively solve the incompatibility of the determination methods for the protein-protein interaction and protein-DNA interaction in the same experimental system in the prior art.
Owner:HUAZHONG AGRI UNIV
Bioactive carbon nanotube composite functionalized with b-sheet polypeptide block copolymer, and preparation method thereof
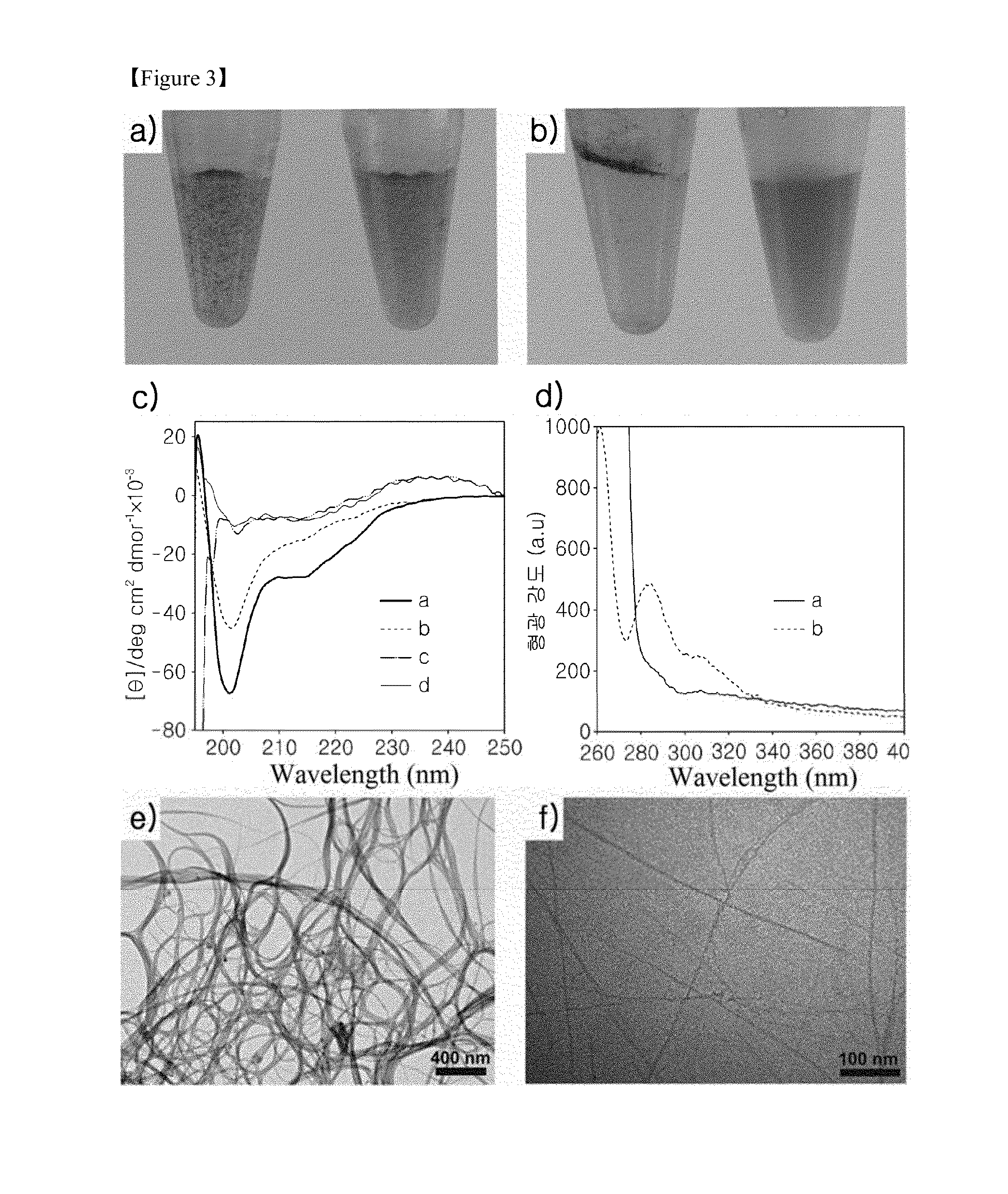
The present invention relates to a bioactive carbon nanotube composite functionalized with a β-sheet polypeptide block copolymer by combination self-assembly, which shows excellent water dispersion, and has biological activity so as to be used as stimulus-responsive and adaptable biomaterials or in the manufacture of CNT-based electronic biosensor devices. In addition, the bioactive carbon nanotube composite can be used as a composition for delivery of a biological active material into cells. Further, the application of the interaction between a β-sheet peptide and a carbon-based hydrophobic material is expected to be useful for designing and developing an inhibitor for diseases caused by the abnormal folding of a protein and by biomacromolecular interactions (protein-protein, protein-DNA, and protein-RNA interactions etc).
Owner:IND ACADEMIC CORP FOUND YONSEI UNIV
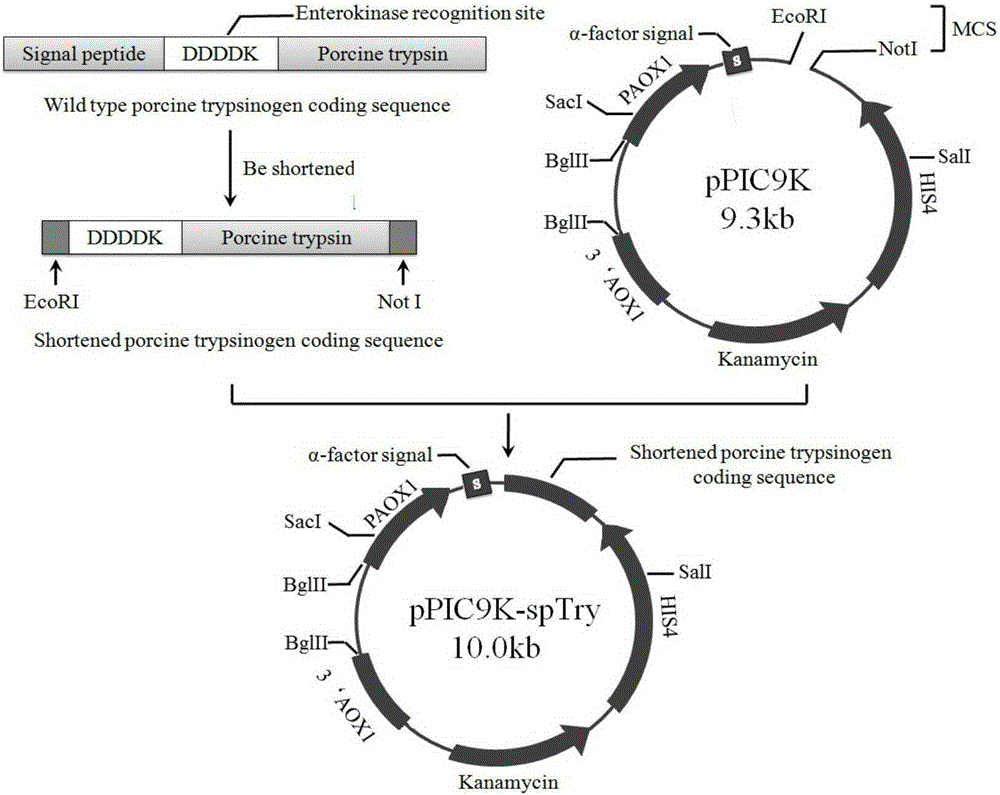
Mutant-type Sus scrofa swine trypsin and encoding gene thereof as well as acquisition method and application
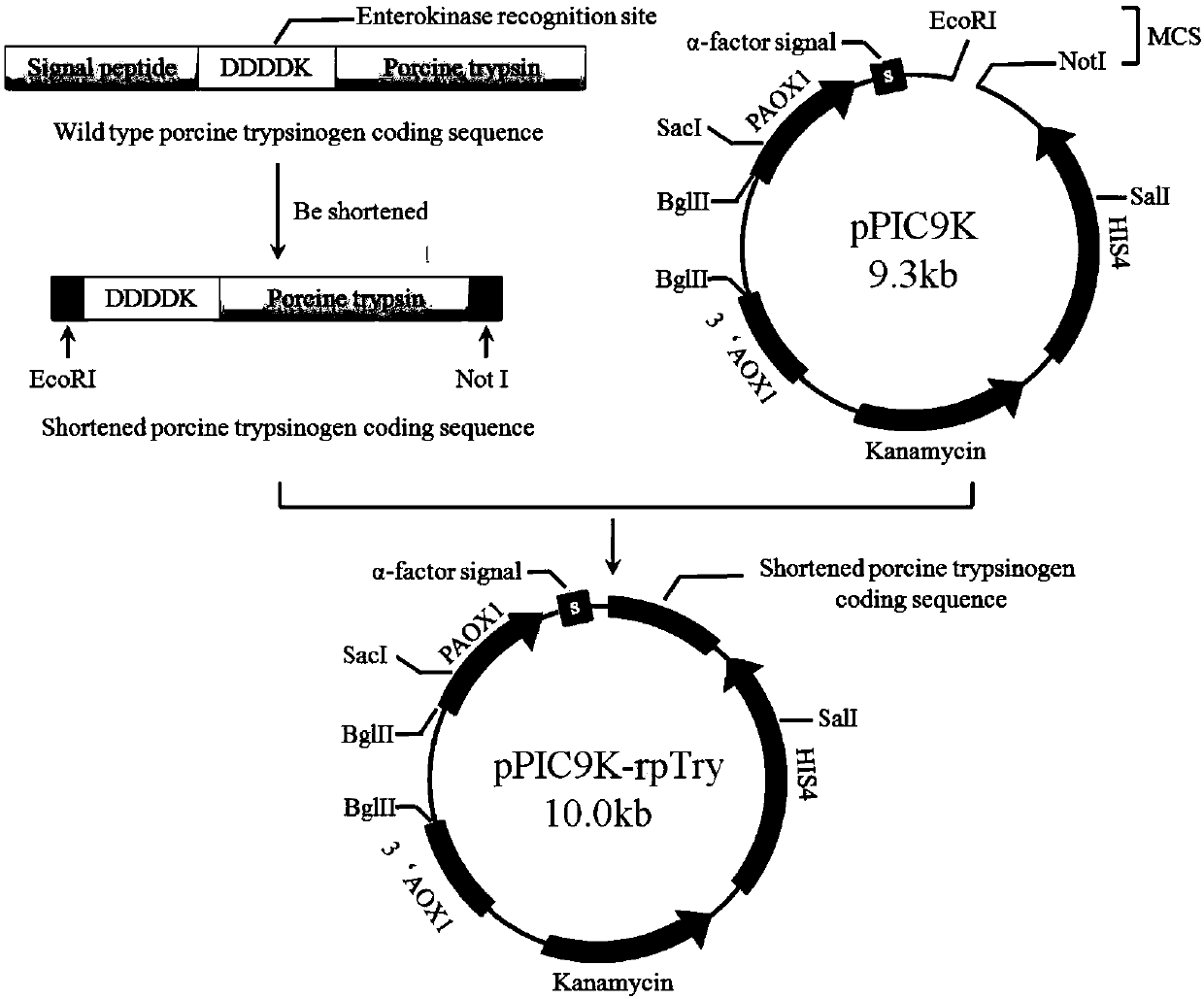
ActiveCN105296452AImprove thermal stabilityClean thoroughlyFungiMicroorganism based processesBiotechnologyWild type
The invention relates to a mutant-type Sus scrofa swine trypsin and an encoding gene thereof as well as an acquisition method and an application. According to the invention, the original wild type Sus scrofa swine trypsin is subjected to mutation to obtain the mutant enzyme with heat stability obviously improved; the invention further provides the encoding gene for encoding the trypsin mutant, the amino acid sequence and a construction method of a trypsin yeast engineering bacteria, wherein the amino acid sequence is SEQ ID NO.1, and the protein DNA sequence in encoding is SEQ ID NO.2. The invention lays a foundation for the research, development and production of the protease through a bioanalysis method, and is beneficial for the early realization of the clean and low-energy biological tanning, and the bio-safe production of leather. With the strain for producing swine trypsin, the yield is relatively high, the process is relatively simple, and the industrial application is facilitated.
Owner:TIANJIN UNIV
Methods and deoxyribonucleic acid for the preparation of tissue factor protein
InactiveUS6994988B1Difficult to identifyIsolation of difficultDepsipeptidesPeptide preparation methodsTissue factorCoagulation Disorder
DNA isolates coding for tissue factor protein and methods of obtaining such DNA and producing tissue factor protein using recombinant expression systems for use in therapeutic composition for the treatment of coagulation disorders.
Owner:GENENTECH INC
Nano particle composition and anti-tumor application thereof
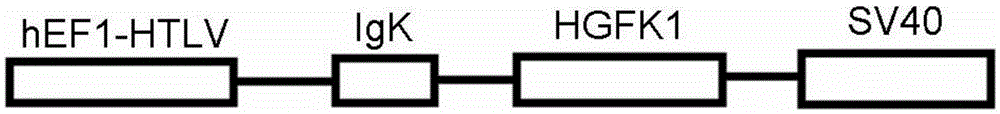
InactiveCN105412943AAvoid diversionPrevent relapsePowder deliveryGenetic material ingredientsTumor targetCyclodextrin
The invention relates to a nano particle composition and anti-tumor application thereof. The nano particle composition is a nano particle formed by assembling a polymer carrier and plasmid for expressing a therapeutic gene; the polymer carrier is a cyclodextrin-polyethyleneimine-polyethylene glycol-folic acid quaternary assembly-type polycation carrier, and the plasmid for expressing the therapeutic gene is eukaryotic expression plasmid containing an HGFK1 protein DNA sequence for coding a hepatocyte growth factor. Compared with the prior art, the nano particle has the advantages of being efficient and tumor targeted; the nano particle can be automatically assembled in an aqueous solution through electrostatic interaction, and the particle size of the nano particle and the stability in a salt solution has an important effect on the function of the nano particle; it is verified that the nano particle composition can prevent transfer or relapse of a cancer and alleviate or relieve or control or improve or cure a primary cancer or a metastatic cancer or other related symptoms and can also prolong the life or survival time of a cancer patient and reduce the mortality.
Owner:XUZHOU MEDICAL COLLEGE
Coumermycin/novobiocin-regulated gene expression system
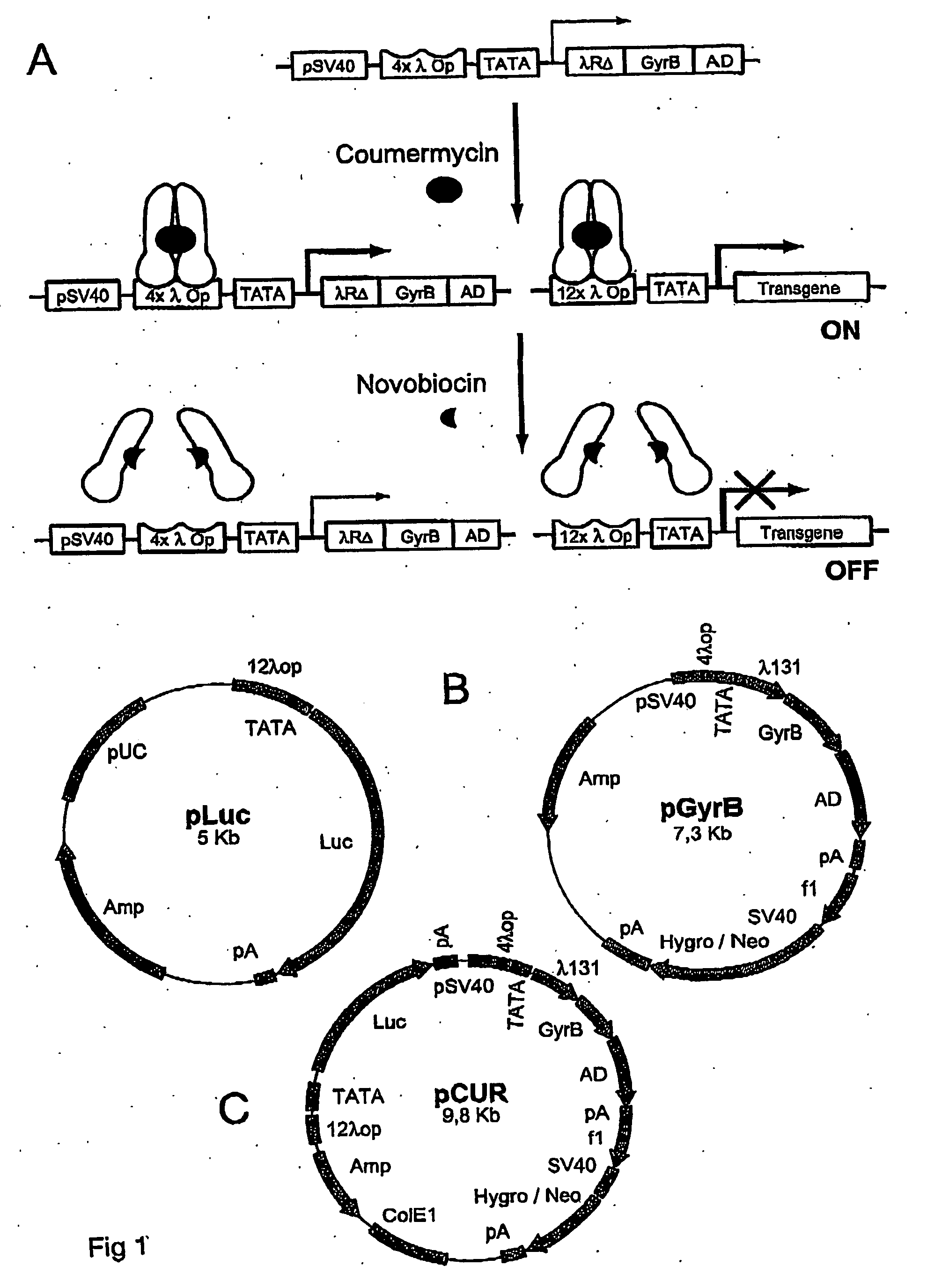
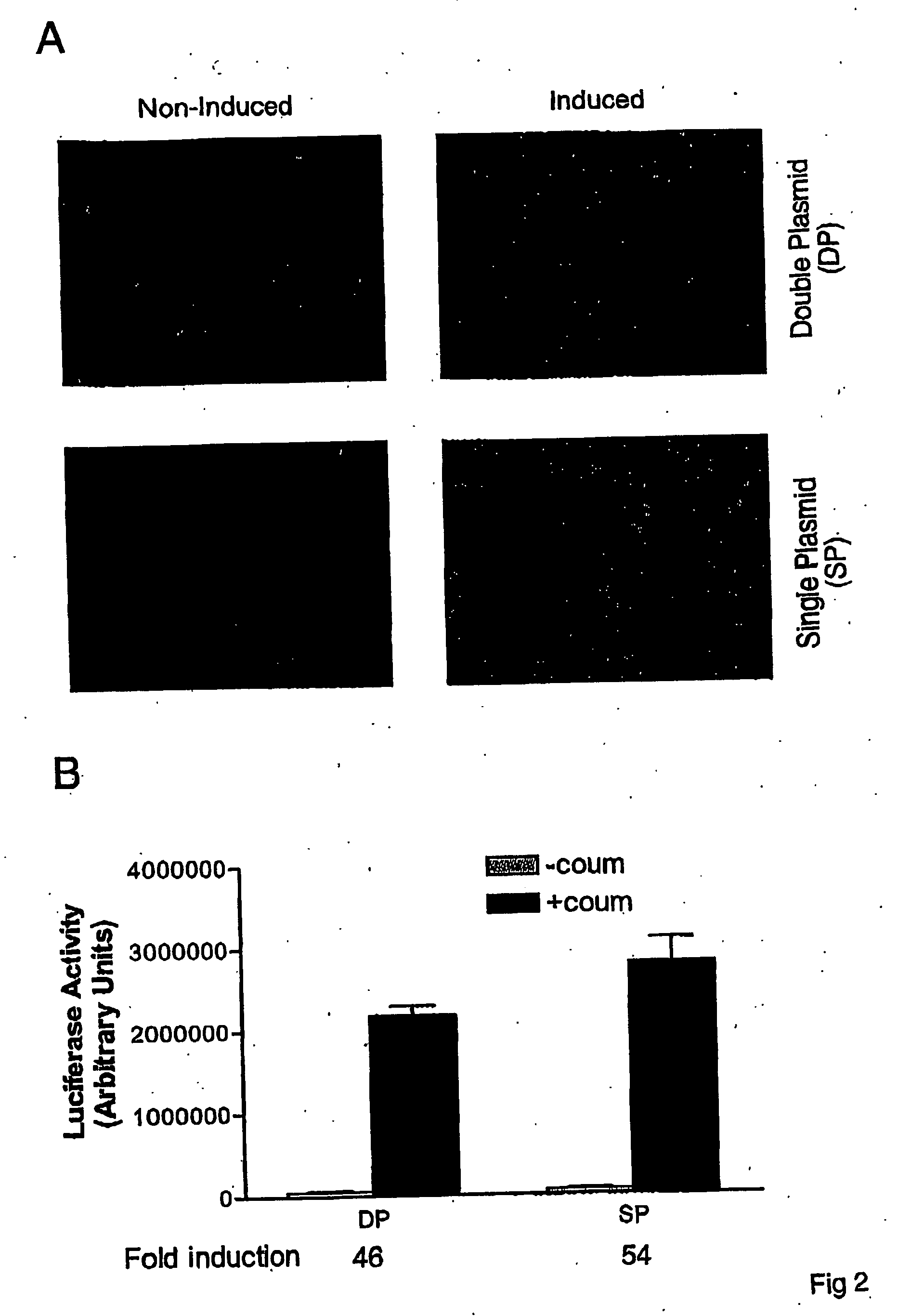
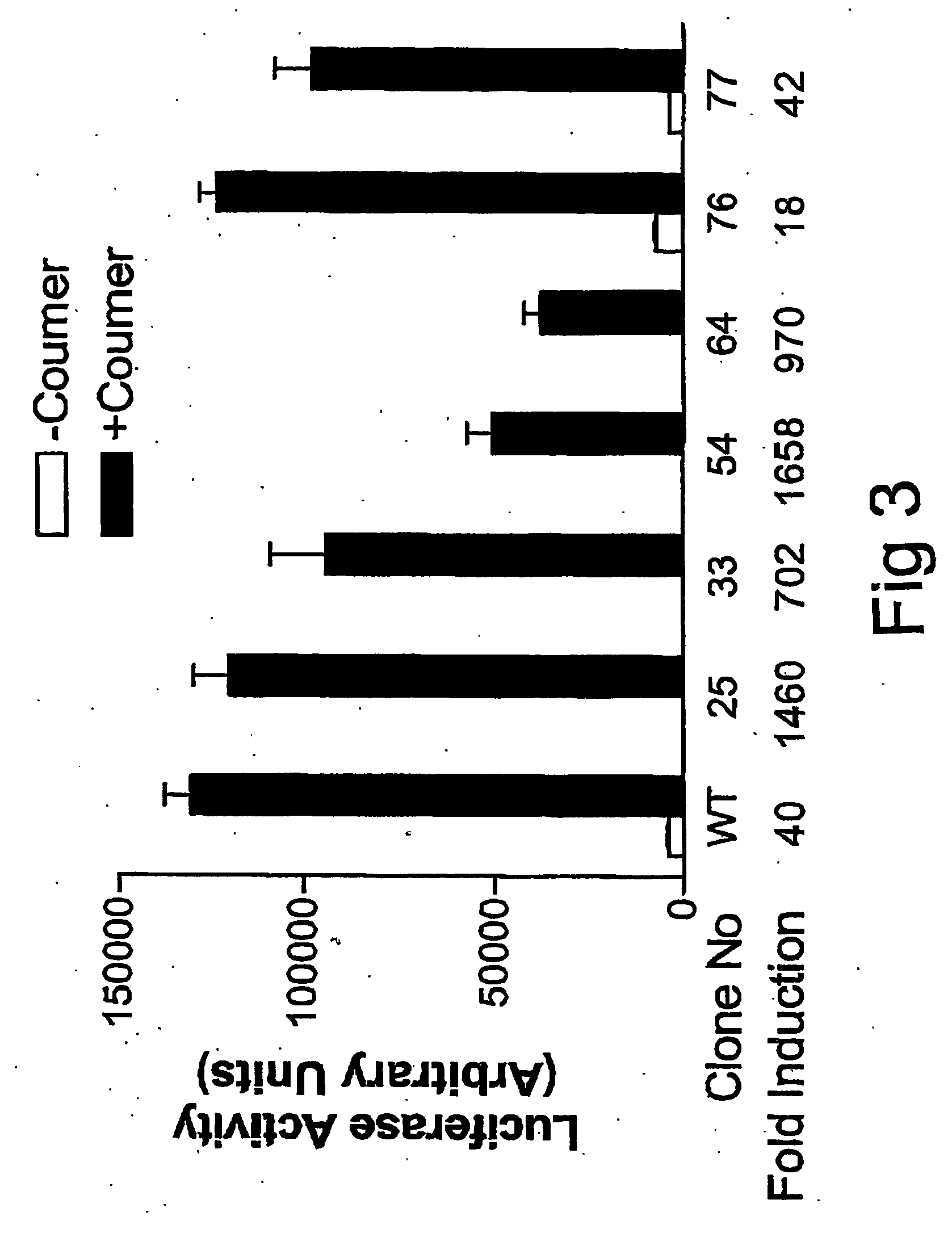
ActiveUS20070026486A1Rapid on/off switching of gene expressionAntibacterial agentsOrganic active ingredientsNovobiocinOperon
A chimeric transactivator comprises a transcription activation domain, a repressor protein DNA binding domain and the bacterial DNA gyrase B subunit. A target gene is operatively linked to operator DNA sequences recognized by the repressor binding domain. The addition of the antibiotic coumermycin results in a coumermycin-switched dimerization of the transactivator, which then binds to operator DNA sequences and activates transcription of the target gene. The addition of novobiocin switches off expression of the target gene by abolishing coumermycin-induced dimerization of the transactivator.
Owner:NAT RES COUNCIL OF CANADA
Fusion protein for seralbumin and interferon
Owner:INST OF BIOENG ACAD OF MILITARY MEDICAL SCI OF THE CHINESE +2
A method for assaying transcriptional regulatory complexes
ActiveCN104774923BImprove stabilityLow efficiencyMicrobiological testing/measurementDna interactionProtein–DNA interaction
The invention relates to a method for determining a transcriptional control complex, and belongs to the field of molecular biology. The method for determining the transcriptional control complex includes the following steps: preparation of a bait DNA sequence, preparation of a DNA sequence for encoding a bar code protein, construction of a bar code protein library, screening of the transcriptional control complex, and in situ adjacent connection capture of the transcriptional control complex, and the identification of the transcriptional control complex. The invention combines cDNA display technology and the adjacent connection technology to establish a novel method for determining the transcriptional control complex; the determination of transcriptional control complex involves simultaneous determination of a variety of interaction effects of protein-protein and protein-DNA; and the method can realize simultaneous determination of a variety of interaction effects of protein-protein and protein-DNA in a same experimental system, and effectively solve the incompatibility of the determination methods for the protein-protein interaction and protein-DNA interaction in the same experimental system in the prior art.
Owner:HUAZHONG AGRI UNIV
Methods and deoxyribonucleic acid for the preparation of tissue factor protein
DNA isolates coding for tissue factor protein and methods of obtaining such DNA and producing tissue factor protein using recombinant expression systems for use in therapeutic composition for the treatment of coagulation disorders.
Owner:GENENTECH INC
Recombinant sheep prion protein oPrP and preparation method and application thereof
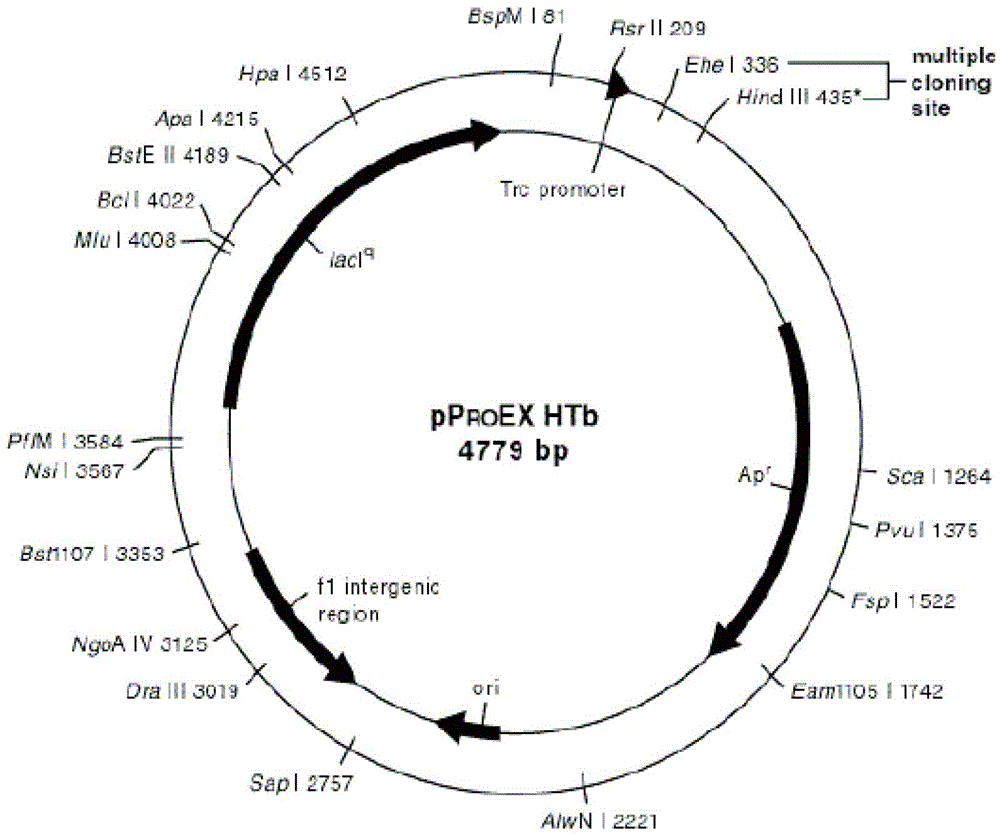
The invention provides a recombinant sheep prion protein oPrP and a preparation method and application thereof. Amino acid sequences of the protein oPrP are shown in SEQ ID No.2. The preparation method includes: using a whole-blood sheep gene as a template, obtaining a sheep prion protein DNA (deoxyribonucleic acid) sequence with enzyme cutting sites by using nucleotide sequences shown in SEQ ID No 3 and 4 as primers and by a PCR (polymerase chain reaction) method, connecting the DNA sequence onto pPROEX-htb plasmids, and removing redundant enzyme cutting sites before a target gene in an expression vector by site-directed mutagenesis, so that an oPrP entire expression vector is obtained; and expressing the vector in escherichia coli, and purifying and renaturing by an overhigh affinity chromatographic column so that the recombinant sheep prion protein is obtained. The amino acid sequences of the obtained recombinant sheep prion protein are highly similar to those of a natural sheep prion protein (only an N terminal comprises one more glycine) and can be used for preparing sheep prion protein monoclonal antibodies and research and development of scrapie diagnostic reagent kits.
Owner:CHINA AGRI UNIV
Antiacid nano oral deoxyribonucleic acid (DNA) anti-tumor vaccine with potential of hydrogen (pH) sensitive characteristic and preparation method
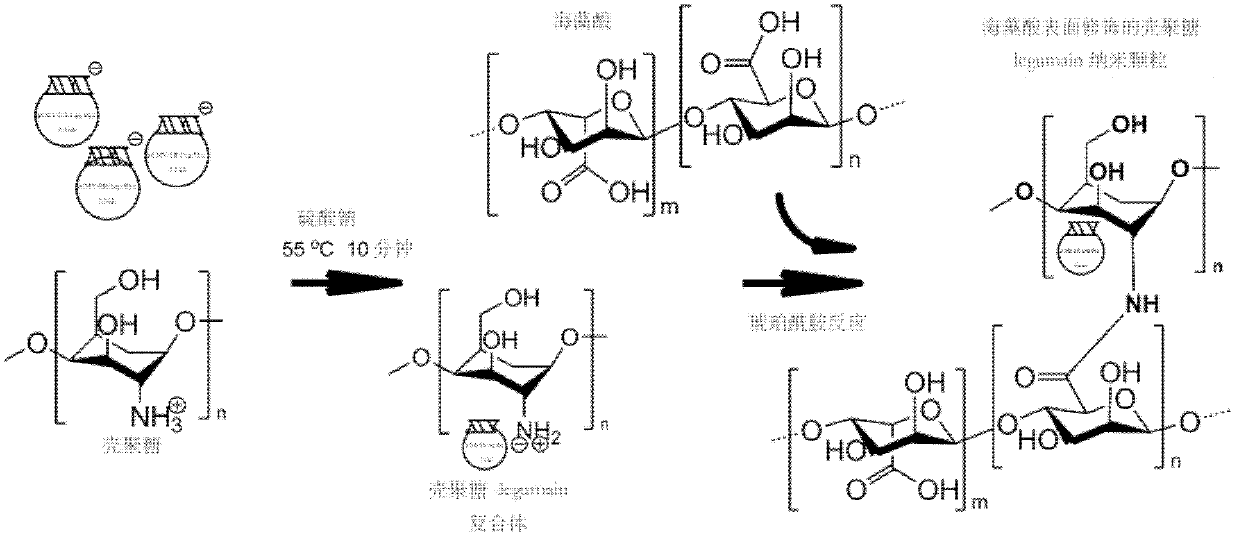
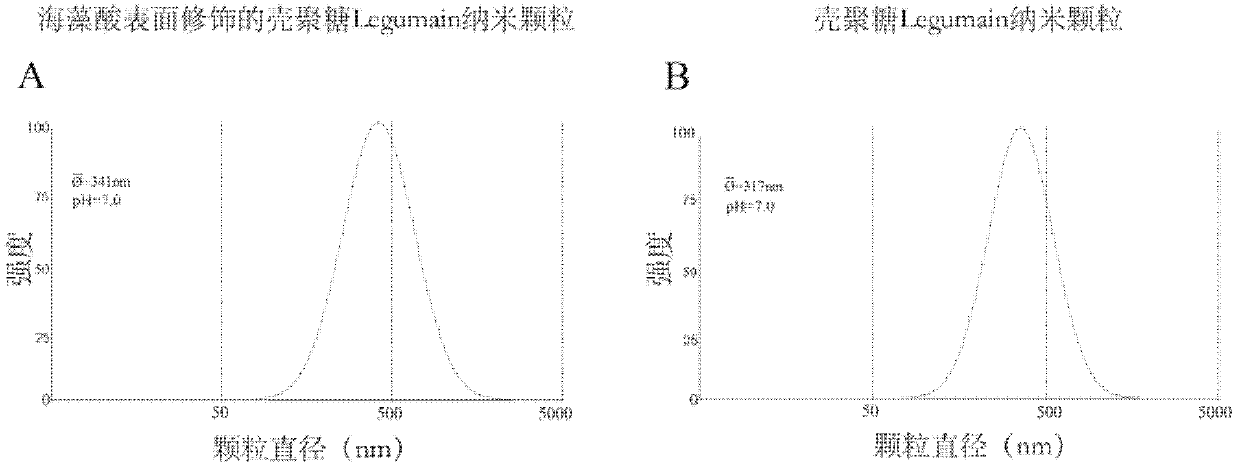
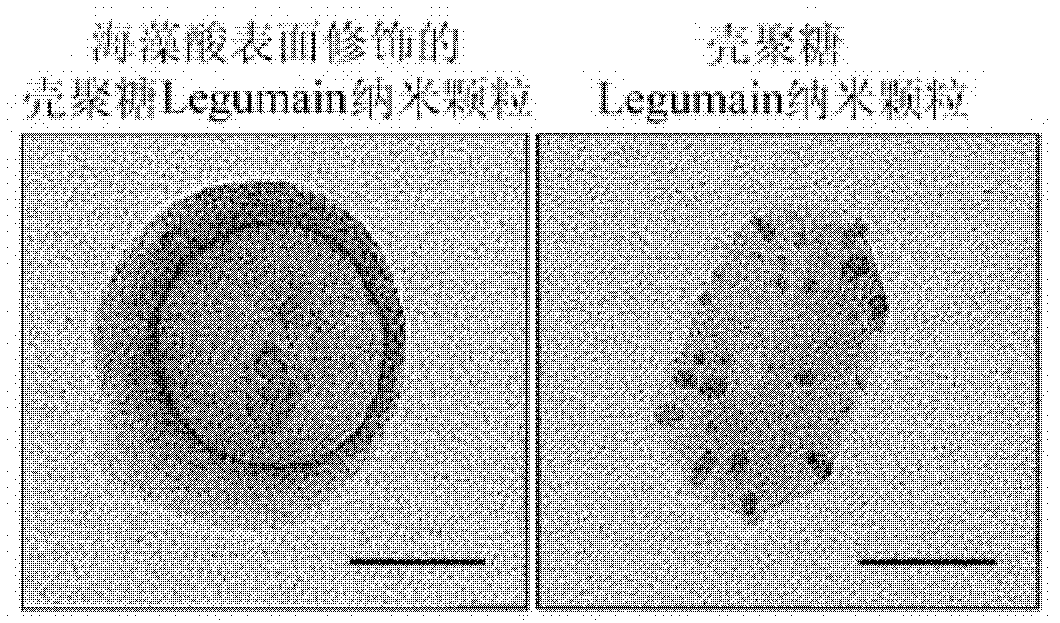
InactiveCN102614527BSmall side effectsStrong antigen presentationGenetic material ingredientsSolution deliveryStainingDendritic cell
Provided are an antiacid nano particle oral deoxyribonucleic acid (DNA) anti-tumor vaccine with the potential of hydrogen (pH) sensitive characteristic and a preparation method. The DNA oral vaccine is nano particles formed by combining chitosan with alginic acid for surface finish and encoded tumor specific antigen Legumain protein DNA plasmids. The DNA oral vaccine is capable of being efficiently phagocytized by dendritic cells and macrophage in peyer's patches and expressing the encoded tumor antigen to activate immune damage of a host for tumor cells. The nano particles have small toxic and side effects and a strong antigen presentation role. Specifically, the DNA plasmids and chitosan are led to be in static negative crosslinking, and then, alginic acid is used for surface finish of chitosan nano particles. By means of immunofluorescent staining and flow cytometry, growth and transfer conditions of breast cancer of a mouse are estimated, immune response of the mouse is analyzed, and antigen presentation capability and anti-tumor effect of the antiacid nano particle oral DNA anti-tumor vaccine with the potential of pH sensitive characteristic are determined.
Owner:NANKAI UNIV
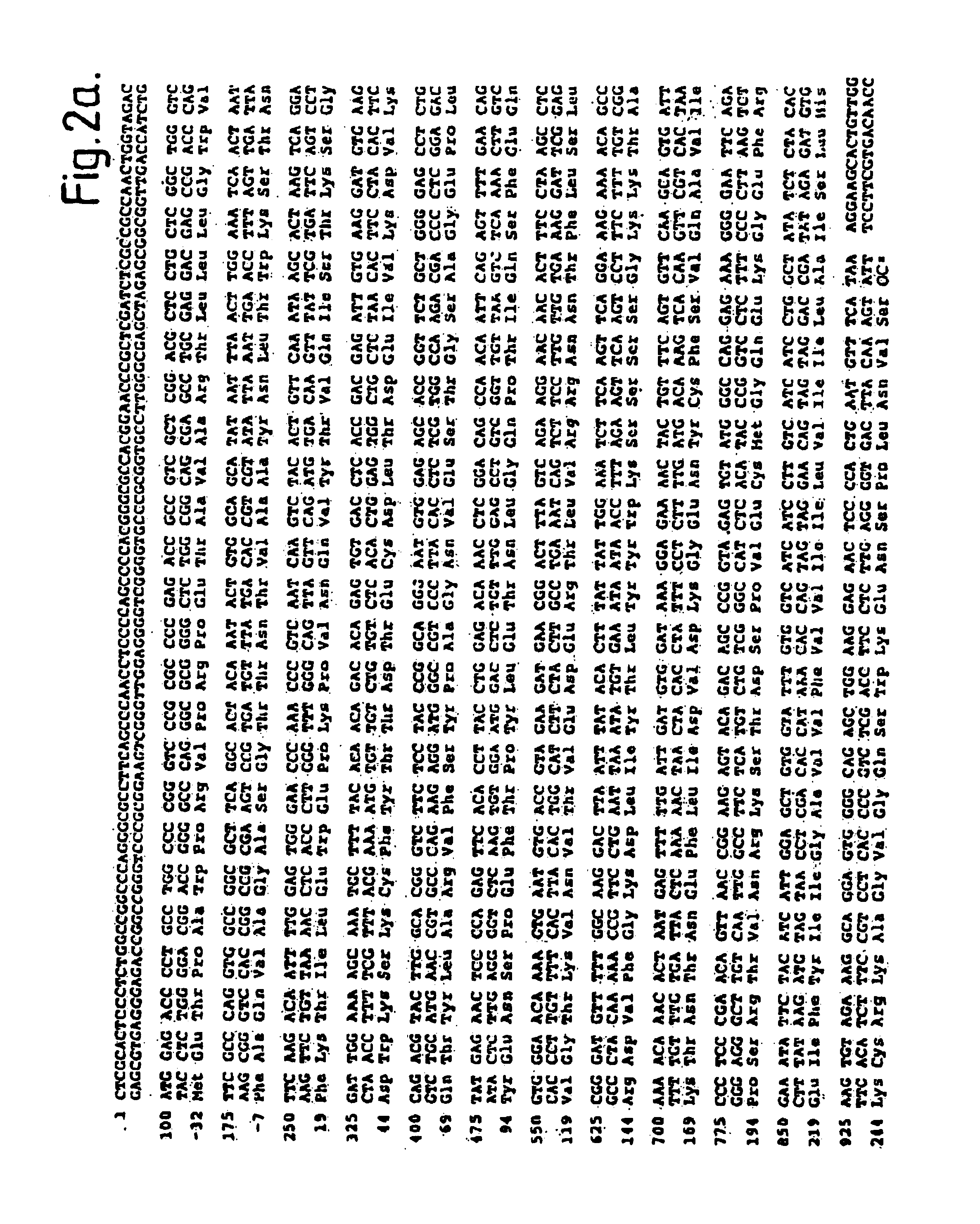
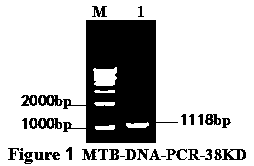
Mycobacterium tuberculosis 38KD protein DNA extraction and recombinant vector construction expressing method
A mycobacterium tuberculosis 38KD protein DNA extraction and recombinant vector construction expressing method comprises the steps of amplification and identification of 38KD protein cDNA, construction of a prokaryotic expression vector MTB38KD, expression and identification of a MTB38KD protein and purification of an 8KD protein.
Owner:AFFILIATED HOSPITAL OF ZUNYI MEDICAL COLLEGE
Preparation and application of human galectin-9 deletant for enhancement of immune response
InactiveCN101792489BImprove the activation effectImprove immunityFungiBacteriaCell immunityImmune inhibition
Owner:INST OF BASIC MEDICAL SCI ACAD OF MILITARY MEDICAL SCI OF PLA
Method for building TALE (transcription activator-like effector) repeated sequences
ActiveCN102787125BHas a cumulative effectShorten the timeMicroorganism based processesGenetic engineeringPlasmid VectorVariome
The invention discloses a side unit used for building TALE (transcription activator-like effector) repeated sequences. The side unit is a repeated unit DNA (deoxyribonucleic acid) segment containing isocaudarner or different flat terminase identification sites at two ends, and the preated unit DNA segment codes repeated units of RVD (repeated variable di-residue) containing NI, NG, HD, NK or NN or variants of the repeated units, wherein in the 5'end isocaudarner or flat terminase identification sites, the 3' end of the identification sites at least has one nucleotide participating in the amino acid coding the N end of the side unit; and in the 3' end isocaudarner or flat terminase identification sites, the 5' end of the identification sites at least has one nucleotide participating in the amino acid coding the C end of the side unit. The side unit has the advantages that the TALE repeated sequences containing any repeated unit number and any ranging sequences, plasmid vectors containing TALE repeated sequences, coding TALE protein DNA combined structural domains and various derived fusion protein plasmid vectors can be conveniently built.
Owner:PEKING UNIV
A kind of mutant type sus scrofa pig-derived trypsin and its coding gene as well as its acquisition method and application
ActiveCN105296452BImprove thermal stabilityClean thoroughlyFungiMicroorganism based processesBiotechnologyWild type
The invention relates to a mutant-type Sus scrofa swine trypsin and an encoding gene thereof as well as an acquisition method and an application. According to the invention, the original wild type Sus scrofa swine trypsin is subjected to mutation to obtain the mutant enzyme with heat stability obviously improved; the invention further provides the encoding gene for encoding the trypsin mutant, the amino acid sequence and a construction method of a trypsin yeast engineering bacteria, wherein the amino acid sequence is SEQ ID NO.1, and the protein DNA sequence in encoding is SEQ ID NO.2. The invention lays a foundation for the research, development and production of the protease through a bioanalysis method, and is beneficial for the early realization of the clean and low-energy biological tanning, and the bio-safe production of leather. With the strain for producing swine trypsin, the yield is relatively high, the process is relatively simple, and the industrial application is facilitated.
Owner:TIANJIN UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com