Method for determining transcriptional control complex
A technology of transcriptional regulation and complex, which is applied to the measurement/testing of microorganisms, biochemical equipment and methods, etc., can solve the problems of simultaneous detection of protein-nucleic acid interaction, inability to fully respond to information, incompatibility, etc., to achieve Alleviation of incompatibility, improvement of measurement efficiency, and large library capacity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Example 1: Solid Phase Screening
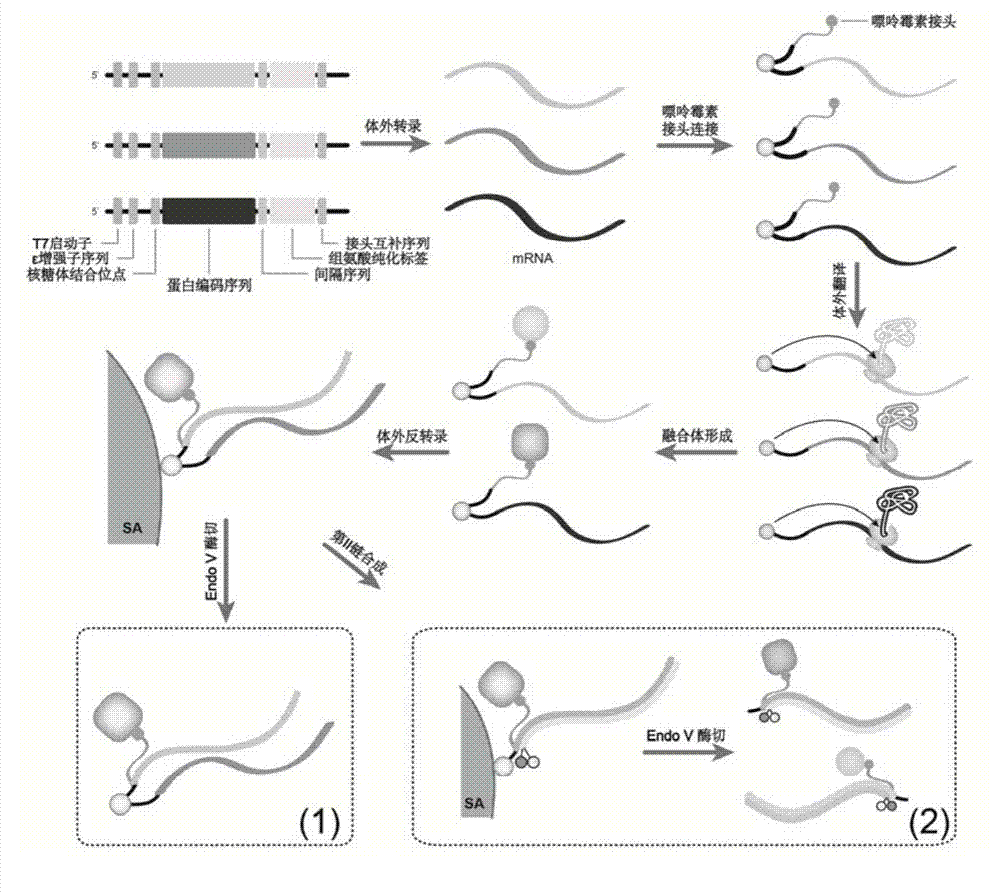
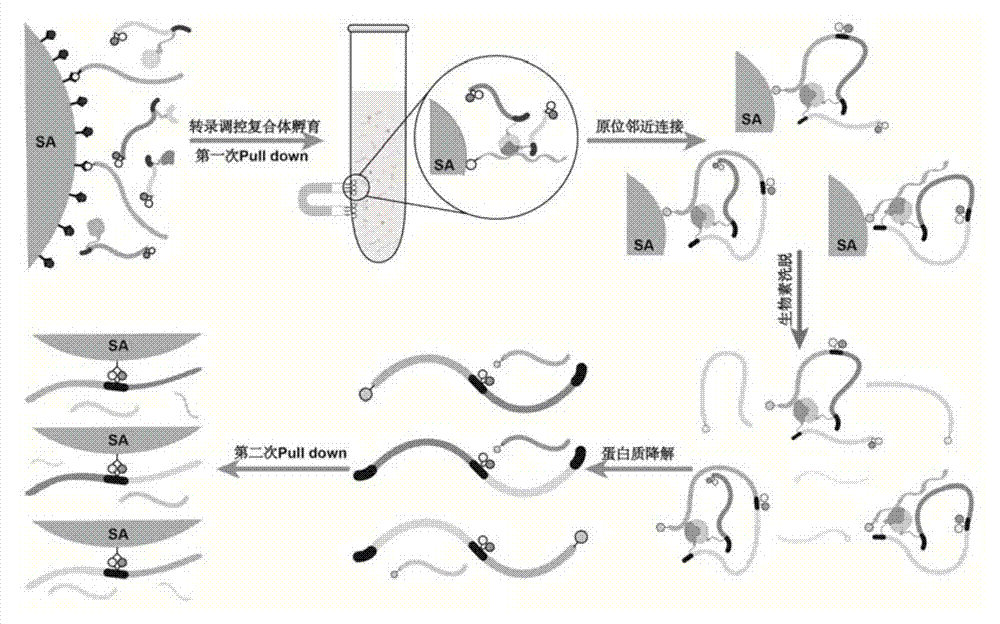
[0072] An embodiment of the method for determining the transcriptional regulatory complex of the present invention, the method for determining the transcriptional regulatory complex described in this embodiment comprises the following steps: 1. Preparation of a bait DNA sequence; 2. Preparation of a DNA sequence encoding a barcode protein; 3. . Construction of barcoded protein library; 4. Solid-phase screening of transcriptional regulatory complexes; 5. In situ proximity ligation capture of transcriptional regulatory complexes; 6. Identification of transcriptional regulatory complexes. Specific steps are as follows:
[0073] 1. Preparation of bait DNA sequence
[0074] Firstly, the double-strand DNA was obtained by overlapping extension PCR. Add 1 μL MDF, 1 μL MDR, 23 μL H 2O, 25 μL premix (Takara), annealing temperature of 60 ° C, 8 cycles, 2% agarose gel detection, PCR products were purified using a High pure PCR product purificat...
Embodiment 2
[0125] Example 2 Liquid Phase Screening
[0126]An embodiment of the method for determining the transcriptional regulatory complex of the present invention, the method for determining the transcriptional regulatory complex described in this embodiment comprises the following steps: 1. Preparation of a bait DNA sequence; 2. Preparation of a DNA sequence encoding a barcode protein; 3. . Construction of barcoded protein library; 4. Liquid-phase screening of transcriptional regulatory complexes; 5. Proximity junction capture of transcriptional regulatory complexes; 6. Identification of transcriptional regulatory complexes. Wherein, steps 1, 2, 3, and 6 are respectively the same as steps 1, 2, 3, and 6 in Example 1 (solid-phase screening), and there are differences in steps 4, 5, and steps 4, 5 are as follows:
[0127] 4. Liquid Phase Screening of Transcription Regulatory Complexes
[0128] 1) Millproe 3K ultrafiltration tube replacement buffer: add 9.6mL pre-cooled transcription ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com