Firefly luciferase and encoding gene and obtaining method thereof
A luciferase and firefly technology, which is applied in the field of encoding genes and the acquisition of the enzyme, can solve the problems of inability to meet the application under high temperature, poor thermal stability and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
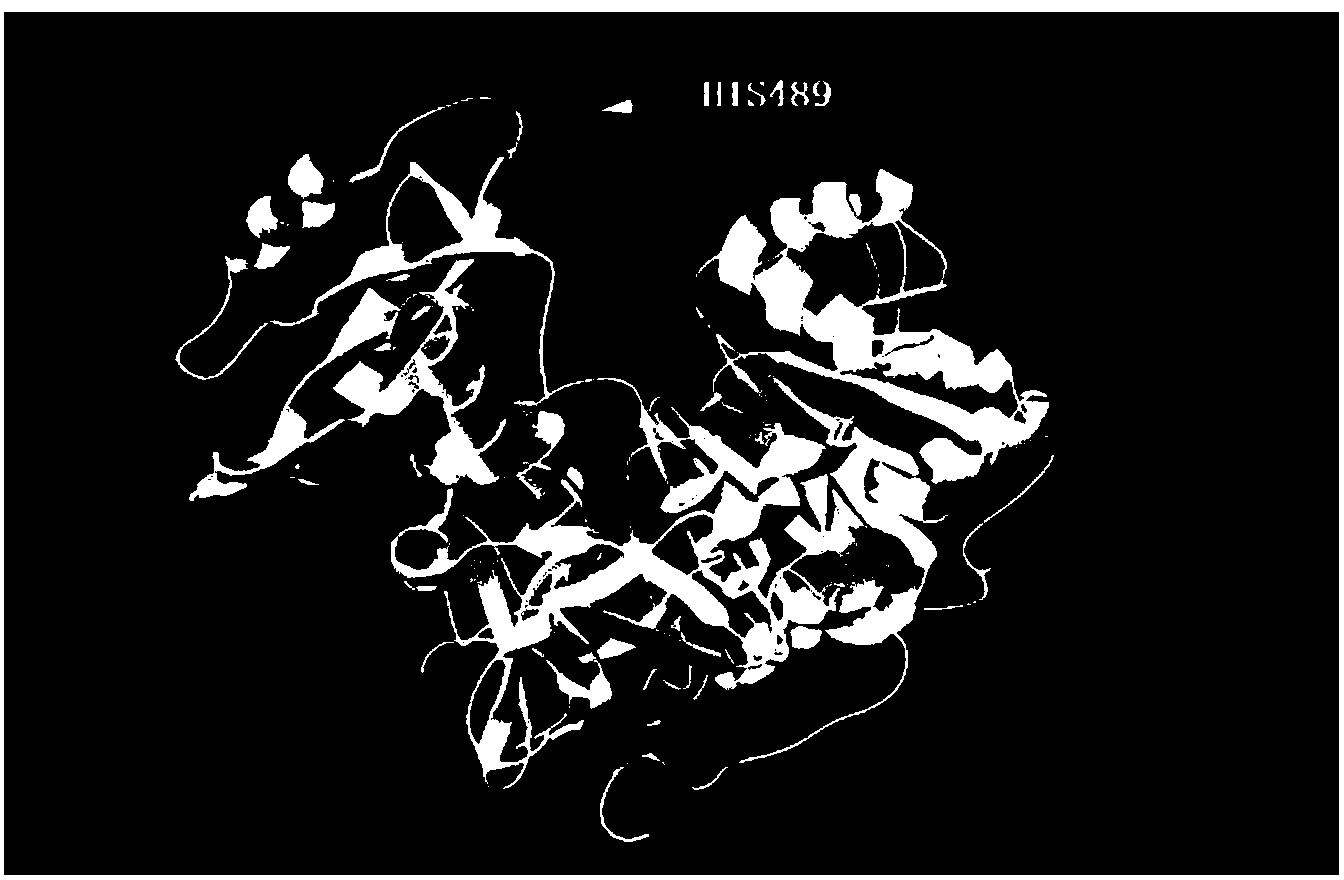
[0054] Embodiment 1 computer-aided design of highly stable mutation site
[0055] Download the coordinate file 1BA3 of luciferase in PDB format from the PDB database (http: / / www.rcsb.org / pdb / home / home.do), and use DeepView4.1 software to process the 1BA3 file: complete missing residues groups and atoms to remove ligand molecules. Perform molecular dynamics simulation on the processed files. All simulations are carried out under the conditions of constant temperature (300K), constant pressure (atmospheric pressure), and force field using Gromos96.1 (53A6), and the final simulation time is 10ns. Select the last 2ns (9-10ns) in the protein molecular dynamics trajectory, and calculate the RMSF (Root Mean Square Fluctuation) value of each amino acid of the protein. This value reflects the conformational status of the amino acid in the molecular dynamics simulation trajectory, and the higher the value, the more unstable the conformation. Through analysis, the region where the acti...
Embodiment 2
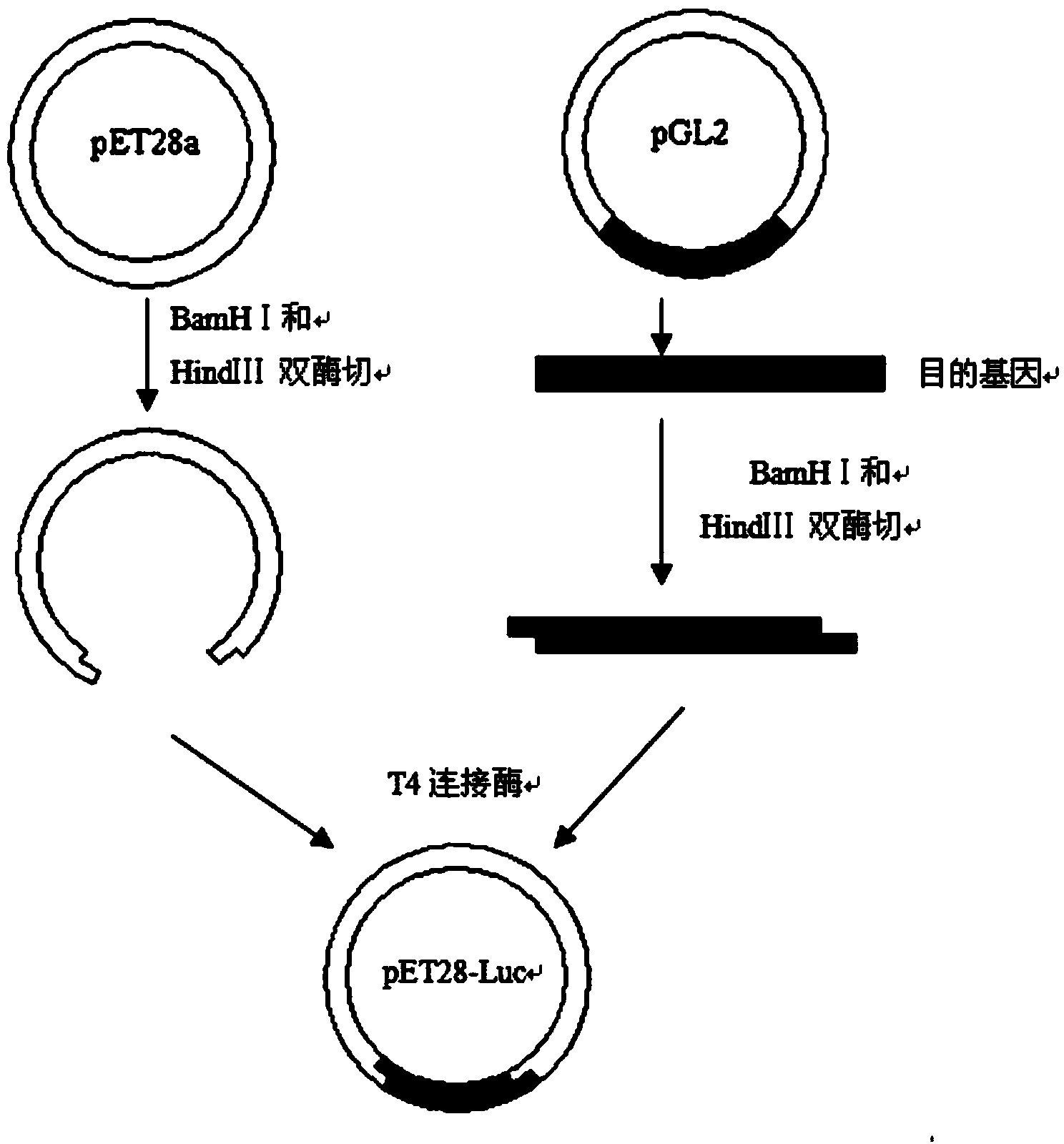
[0056] Embodiment 2 contains the construction of firefly luciferase gene expression vector
[0057] 1. Primer Design
[0058] Using the nucleotide sequence of the plasmid pGL2-control vector (purchased from Promega) containing the North American firefly (P. Design primers for endonuclease sites, as follows:
[0059] Primer luc+:5-CGGGATCCATGGAAGACGCCAAAAAC-3
[0060] Primer luc-:5-CCCAAGCTTTTACAATTTGGACTTTCCGC-3
[0061] In the upstream primer, CG is the protective base, in the upstream primer GGATCC is the restriction site of BamHI, in the downstream primer CCC is the protective base, and AAGCTT is the Hind III endonuclease site. After the properties and parameters of the primers were qualified by oligo6 software, Beijing Aoke Dingsheng Biotechnology Co., Ltd. was entrusted to synthesize the primers.
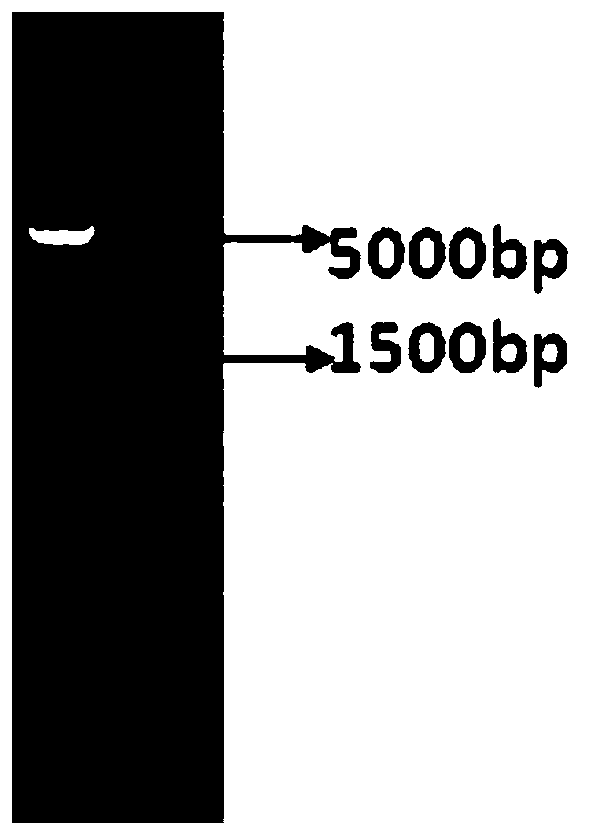
[0062] 2. PCR Amplification of Firefly Luciferase Gene (Luc)
[0063] The plasmid pGL2-control vector (purchased from Promega) containing the Luc gene was used as a templa...
Embodiment 3Q
[0070] Example 3 Quick Change method site-directed mutagenesis of firefly luciferase gene (Luc)
[0071] Rapid site-directed mutagenesis is to directly introduce mutations to amino acids in specific double-stranded DNA by PCR. It is completed in one step and does not require subcloning. This is currently the simplest site-directed mutagenesis method. The principle is: a circular plasmid is used as a template, and a pair of primers containing mutation points are used to amplify. The product is a complete unmethylated plasmid. Because the template plasmid is a methylated plasmid extracted from E. coli, it can The DpnⅠ enzyme digestion method was used to remove the template, and only the newly amplified mutant plasmid remained in the final product. The product could be directly transformed into competent cells of Escherichia coli, and positive clones were selected for sequencing verification. The process is as Figure 4 shown.
[0072] 1. Primer Design
[0073] The mutant primer...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com