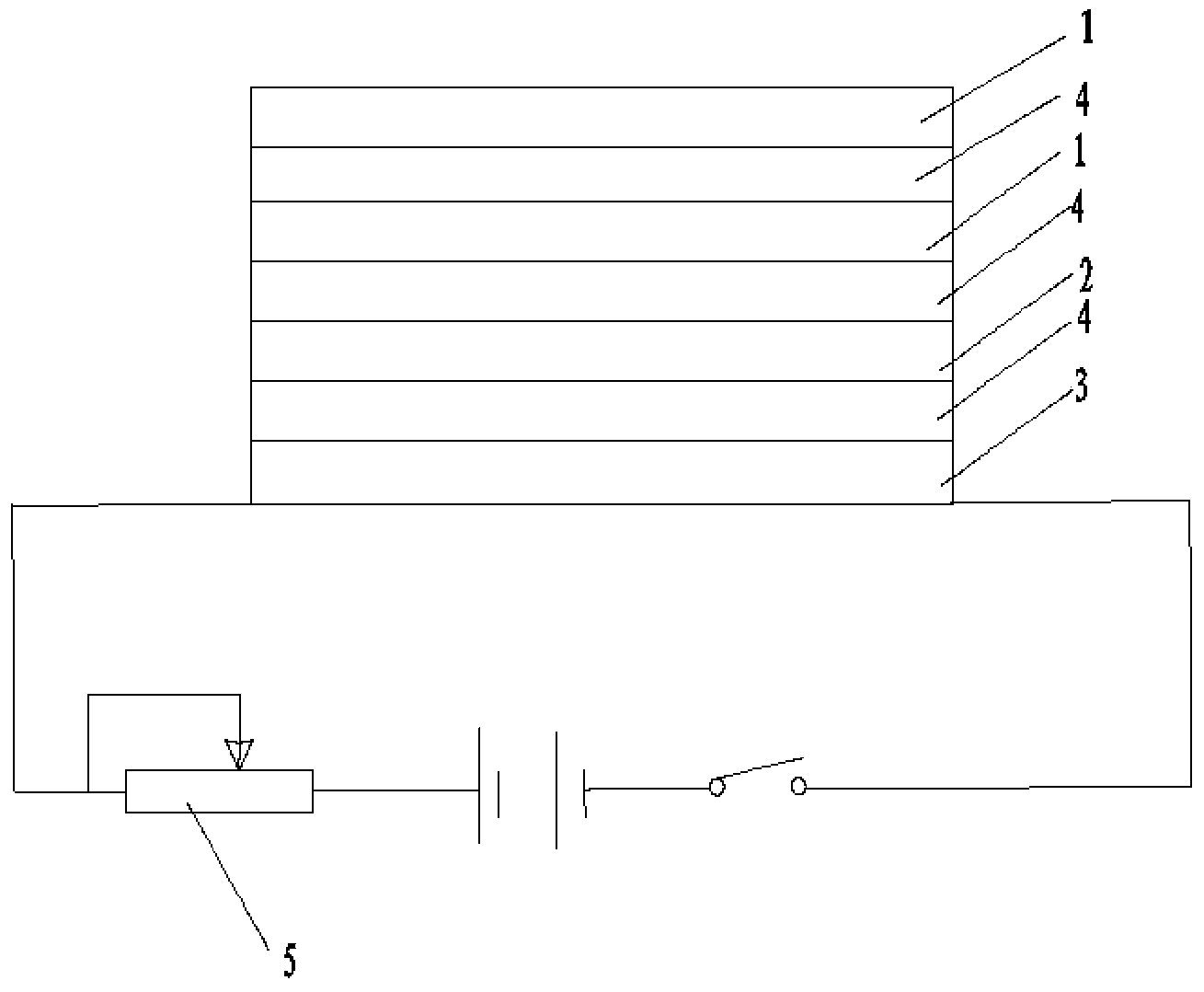
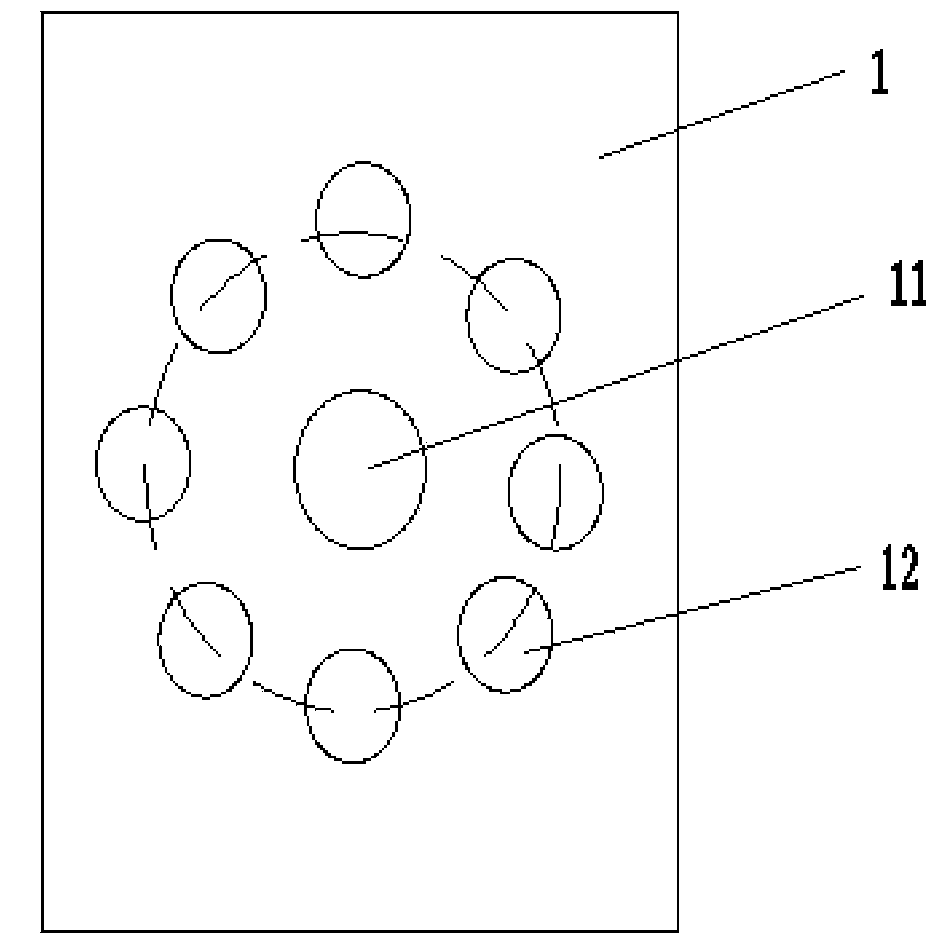
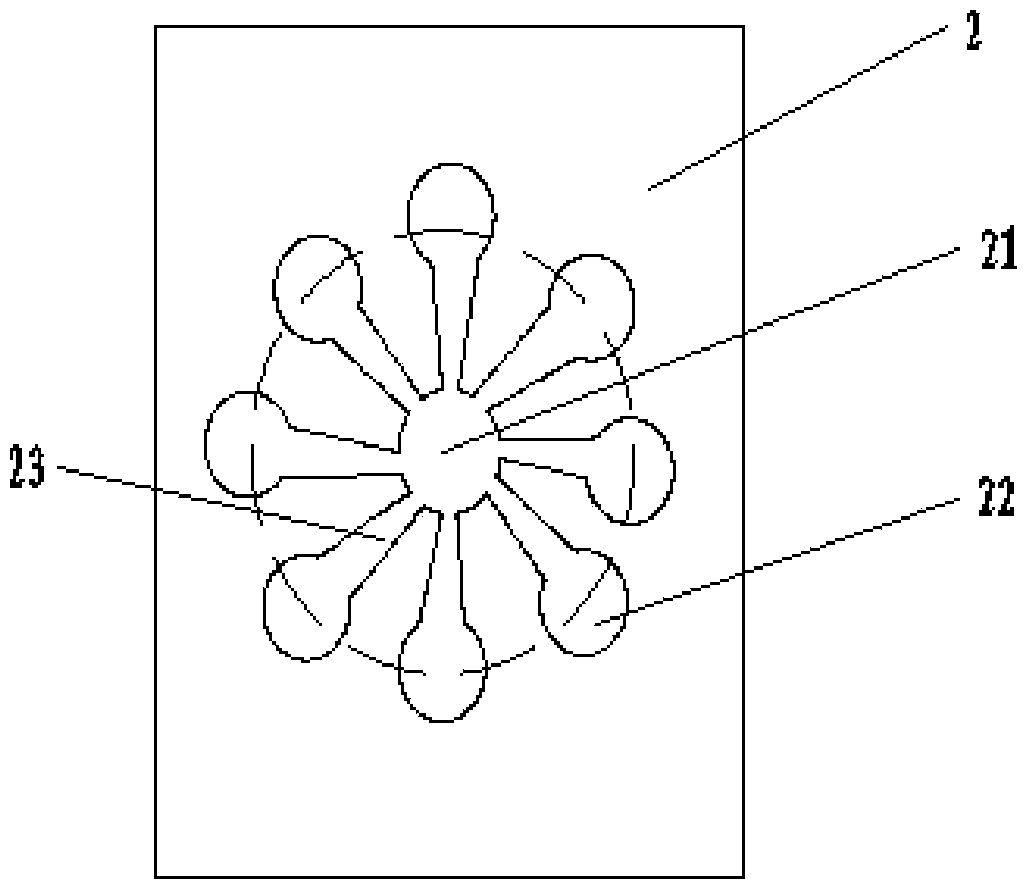
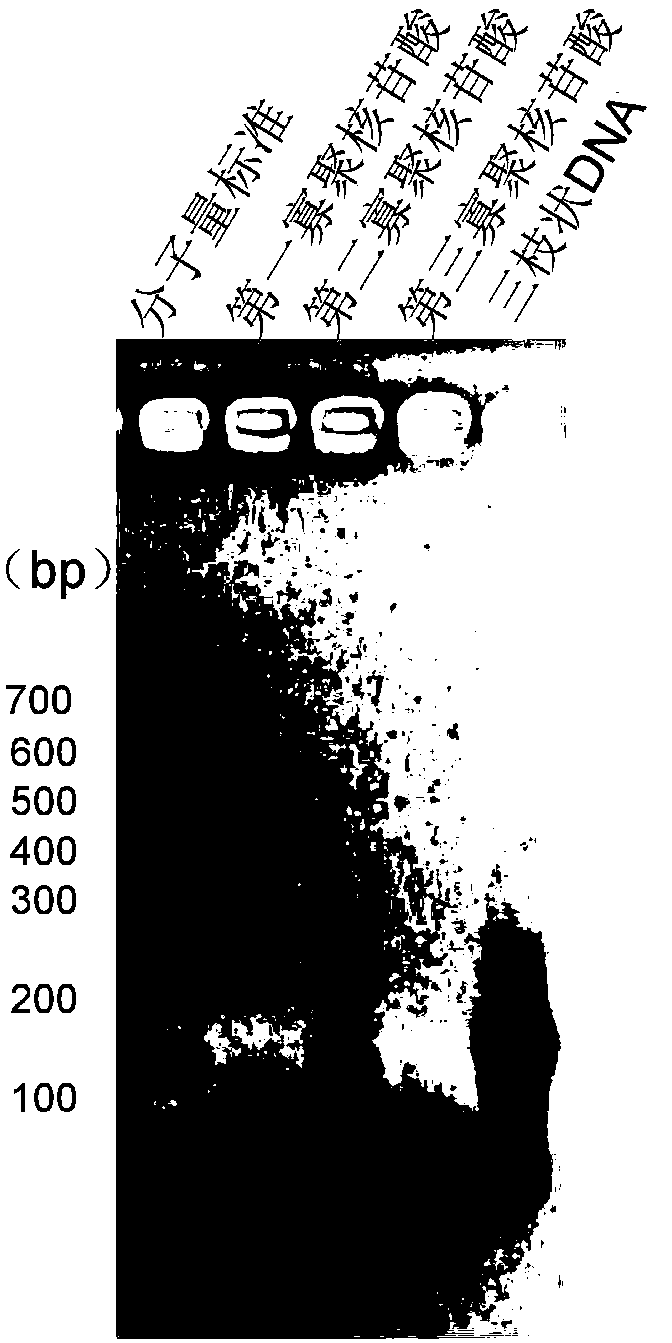
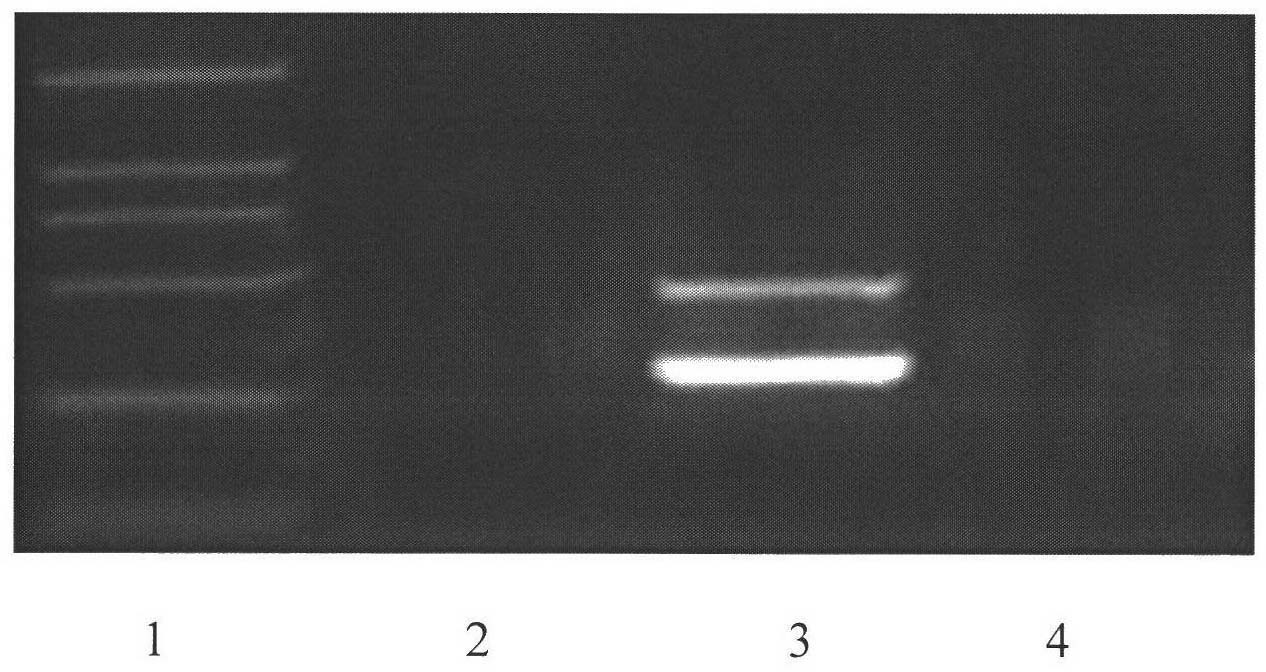
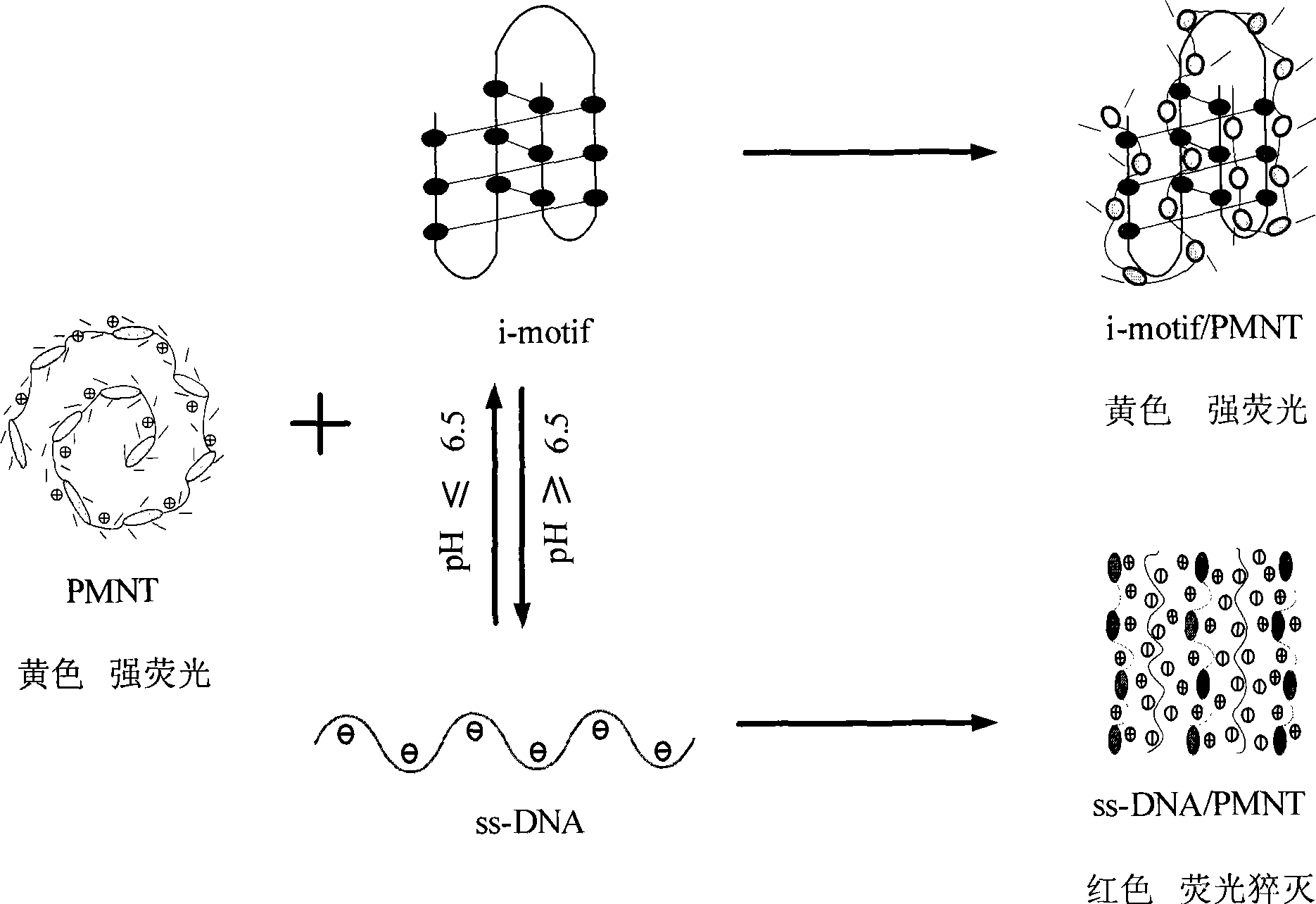
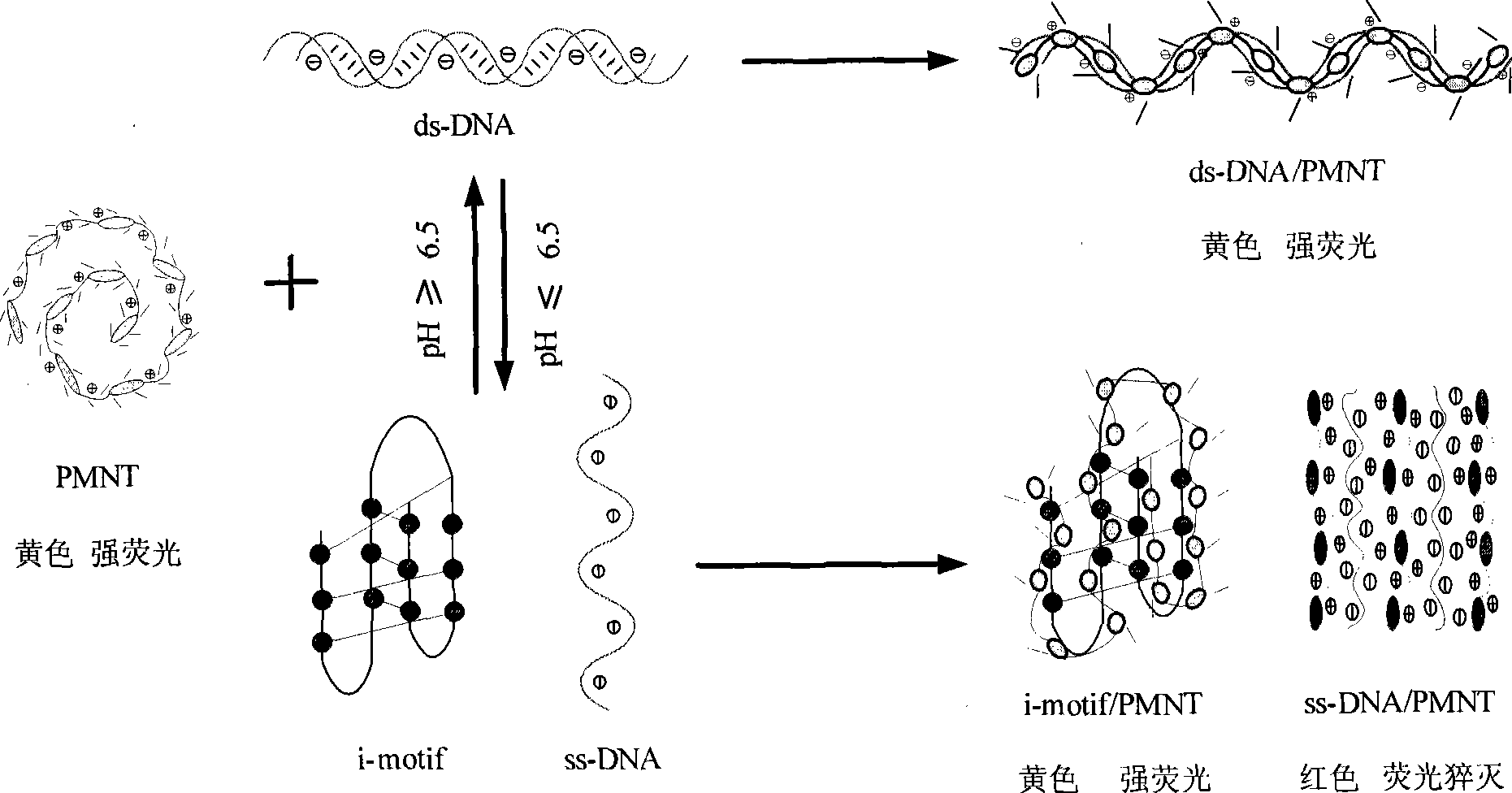
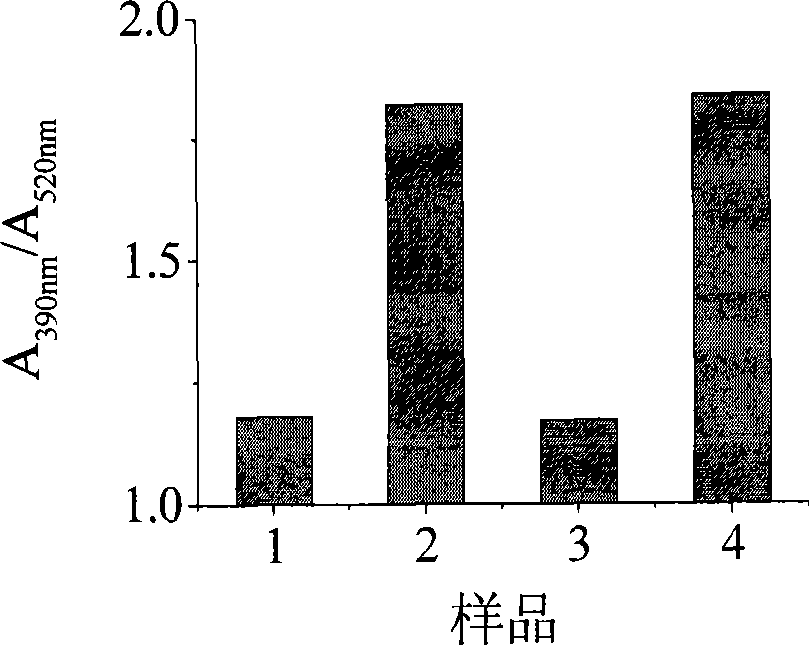
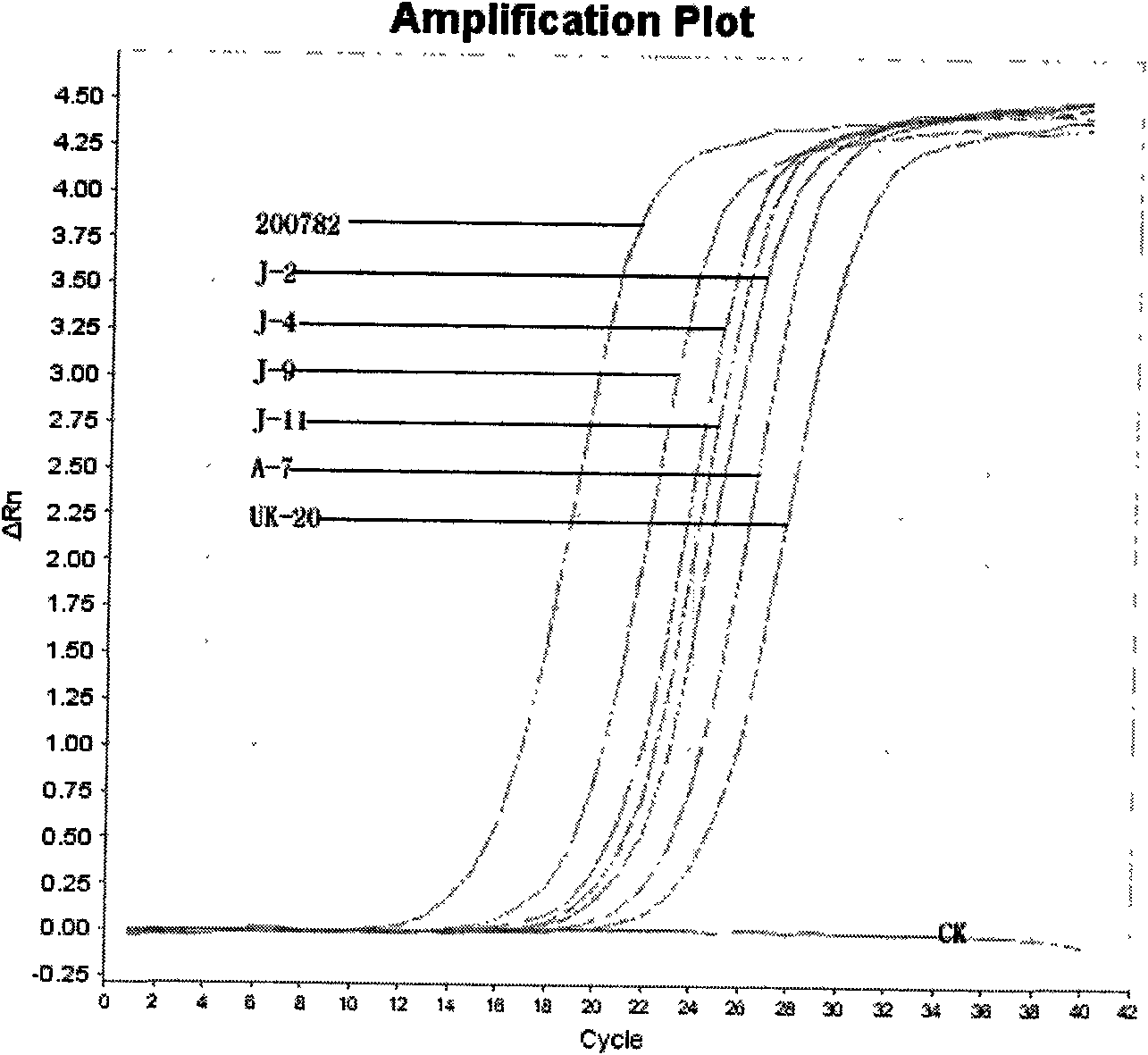
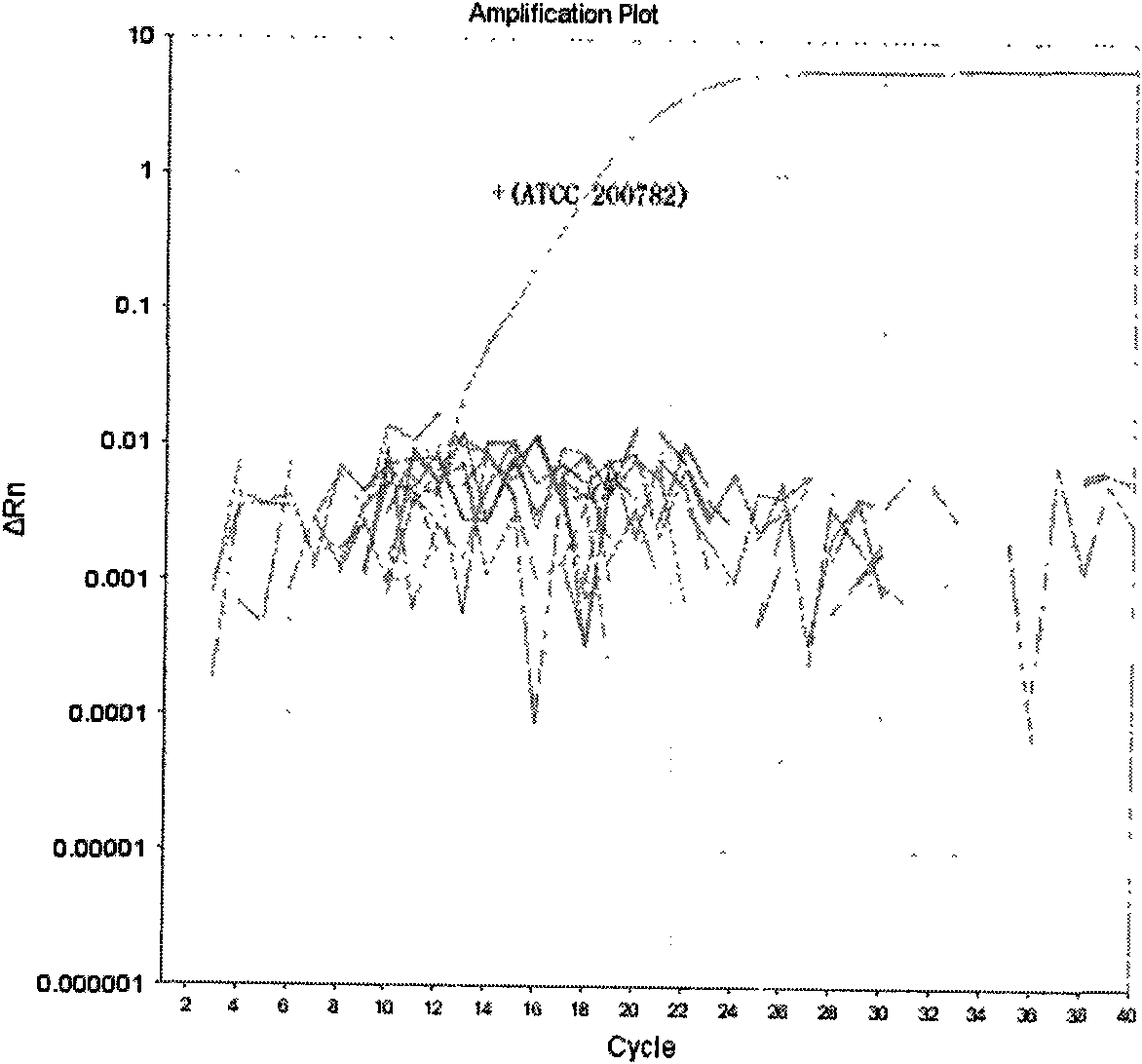
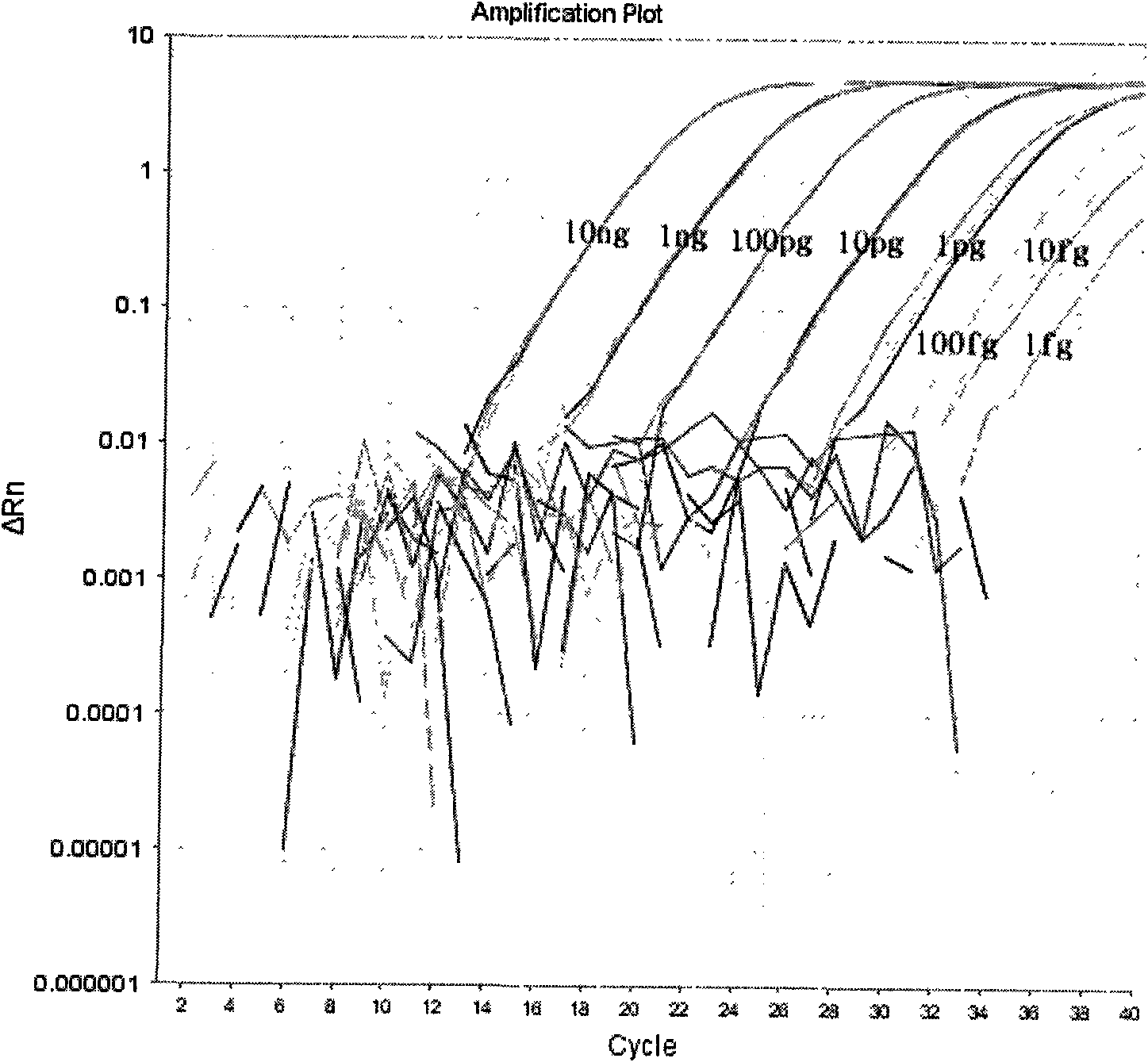
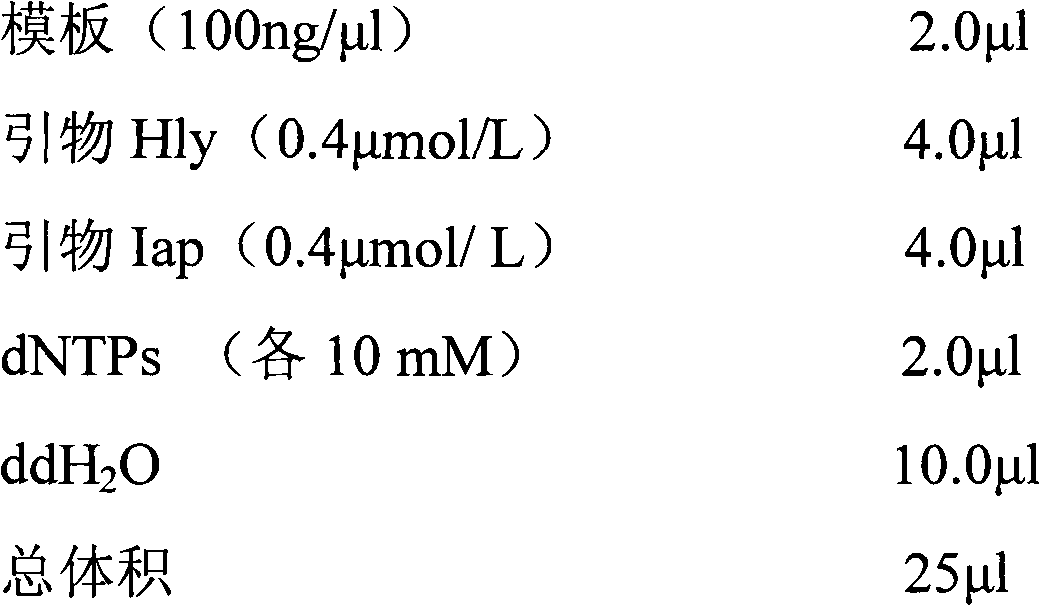Patents
Literature
197 results about "DNA Solutions" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
DNA Solutions is a DNA testing company created by biotechnologist Vern Muir B.Sc. in 1996, and incorporated in 1998. The company created a home paternity kit in 1997 and has since expanded its services to include DNA sample storage.
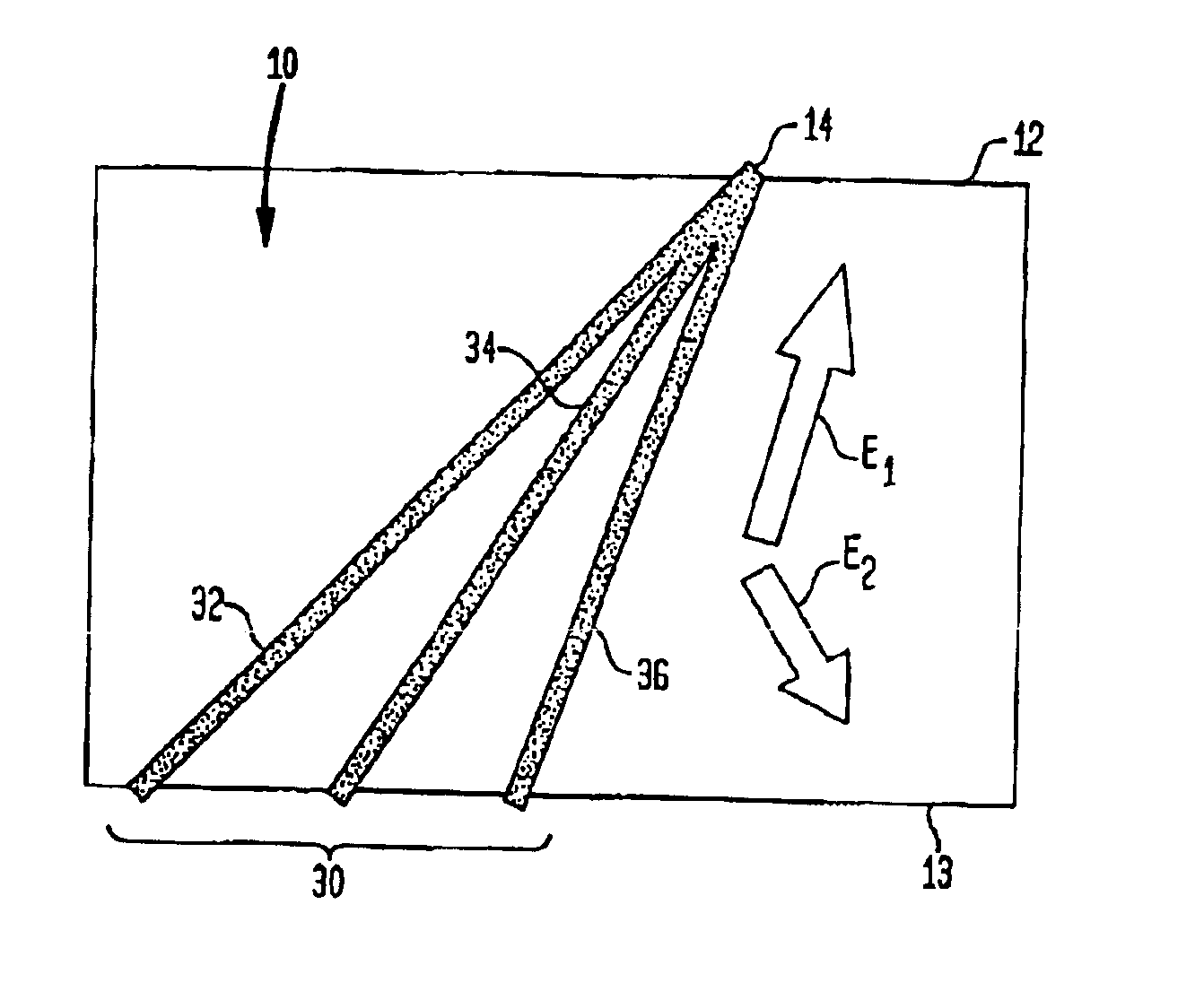
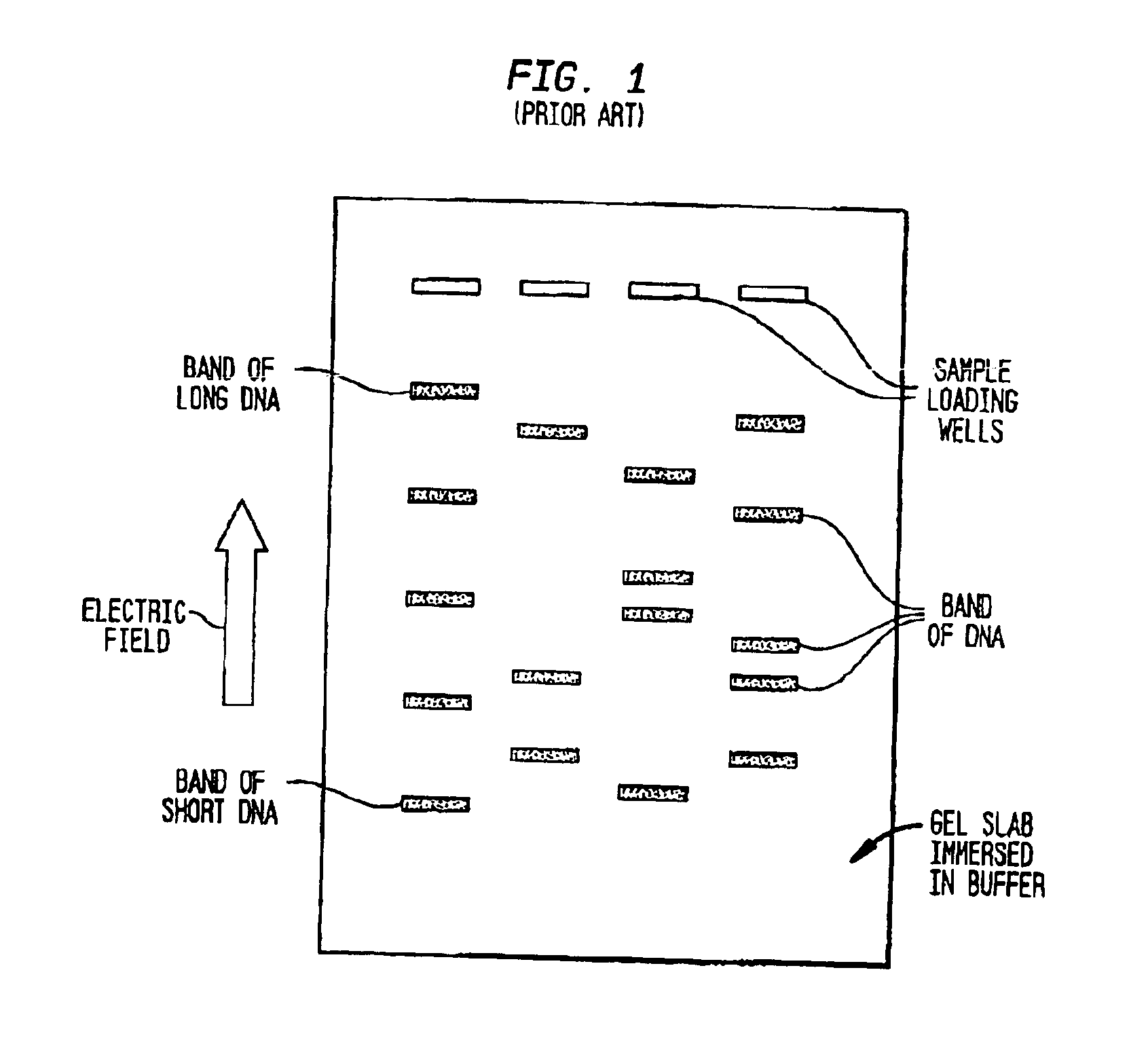
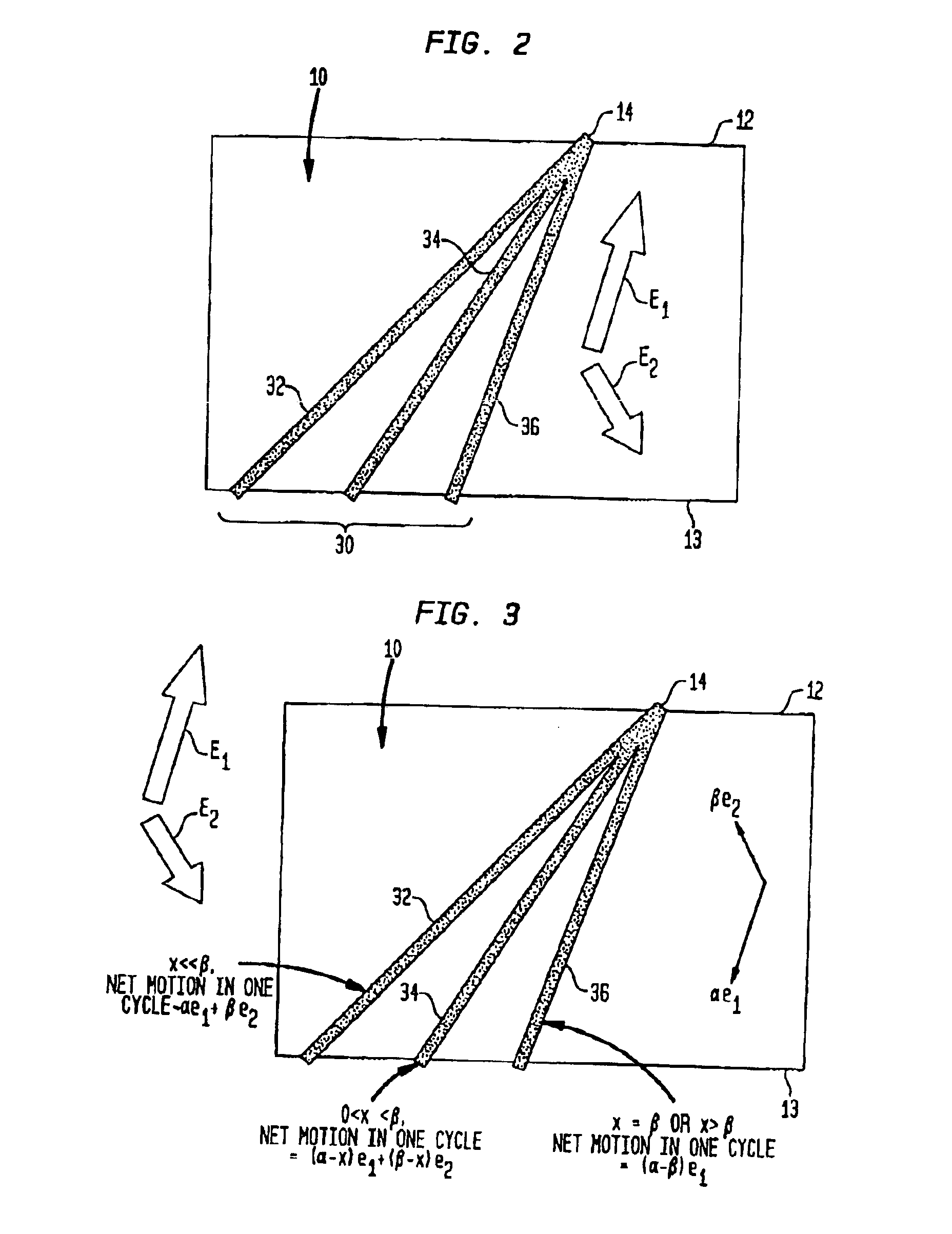
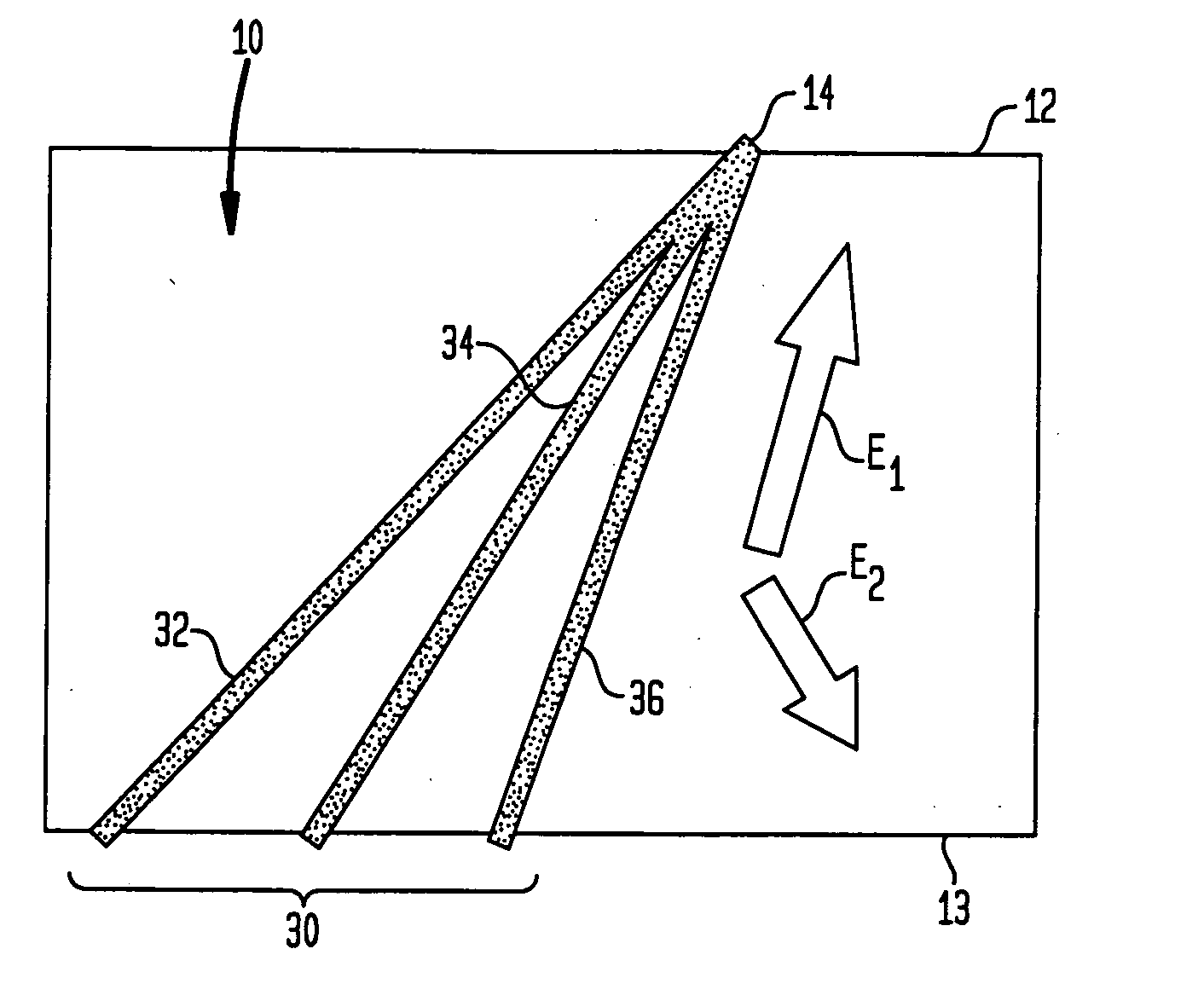
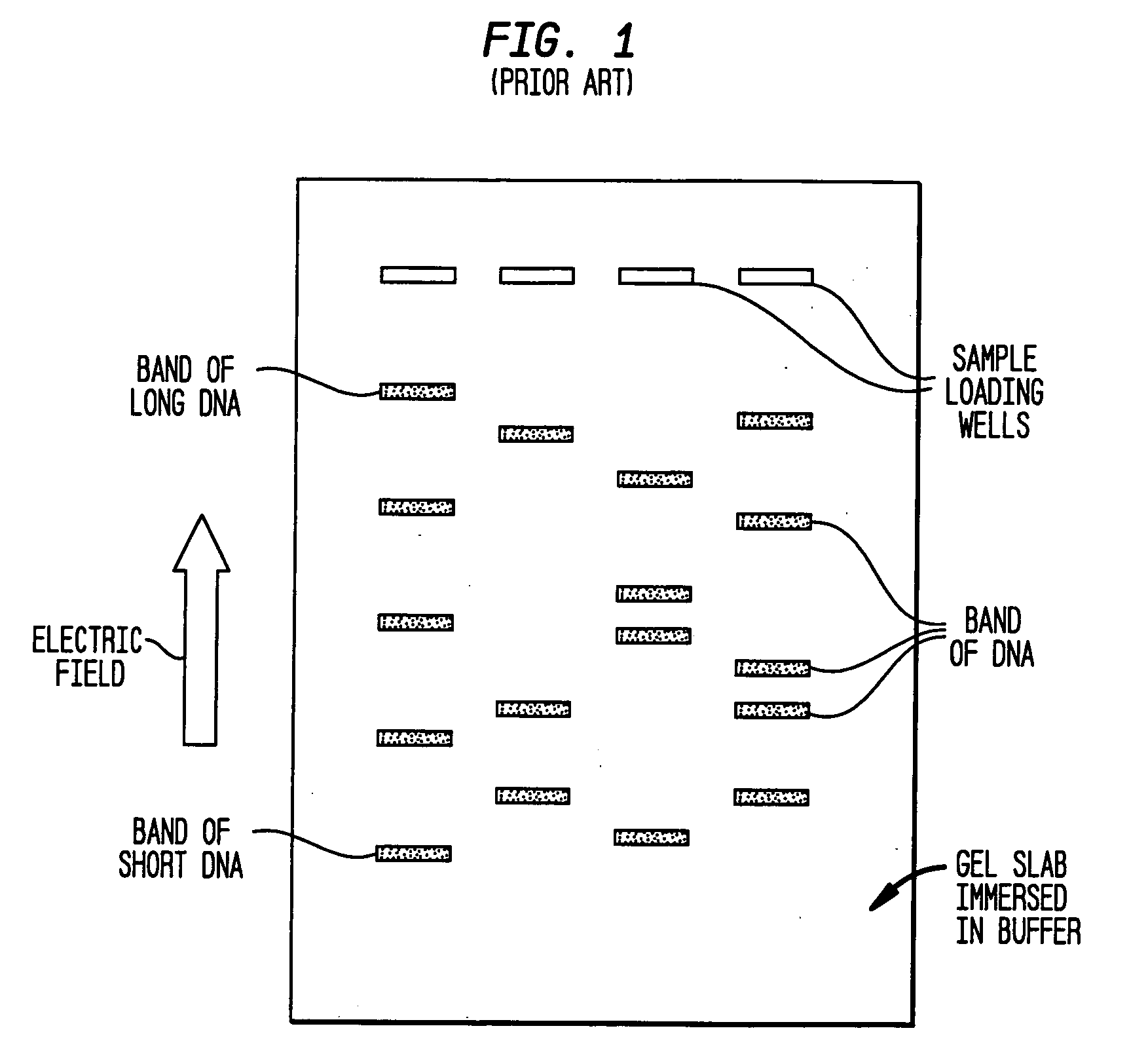
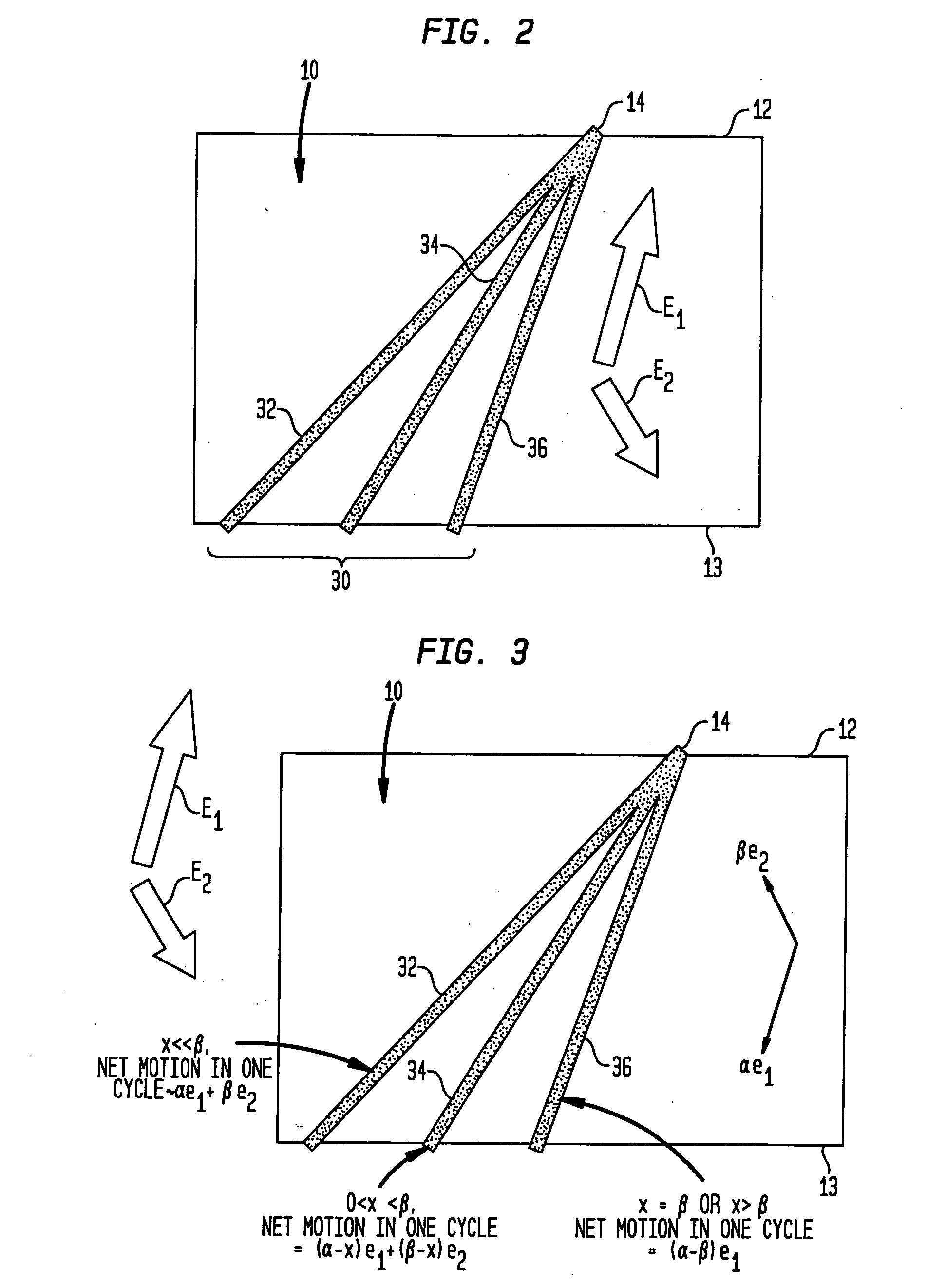
Fractionation of macro-molecules using asymmetric pulsed field electrophoresis
InactiveUS6881317B2Rapid fractionationSludge treatmentVolume/mass flow measurementDNA SolutionsPulsed field electrophoresis
A method and apparatus for fractionation of charged macro-molecules such as DNA is provided. DNA solution is loaded into a matrix including an array of obstacles. An alternating electric field having two different fields at different orientations is applied. The alternating electric field is asymmetric in that one field is stronger in duration or intensity than the other field, or is otherwise asymmetric. The DNA molecules are thereby fractionated according to site and are driven to a far side of the matrix where the fractionated DNA is recovered. The fractionating electric field can be used to load and recover the DNA to operate the process continuously.
Owner:THE TRUSTEES FOR PRINCETON UNIV
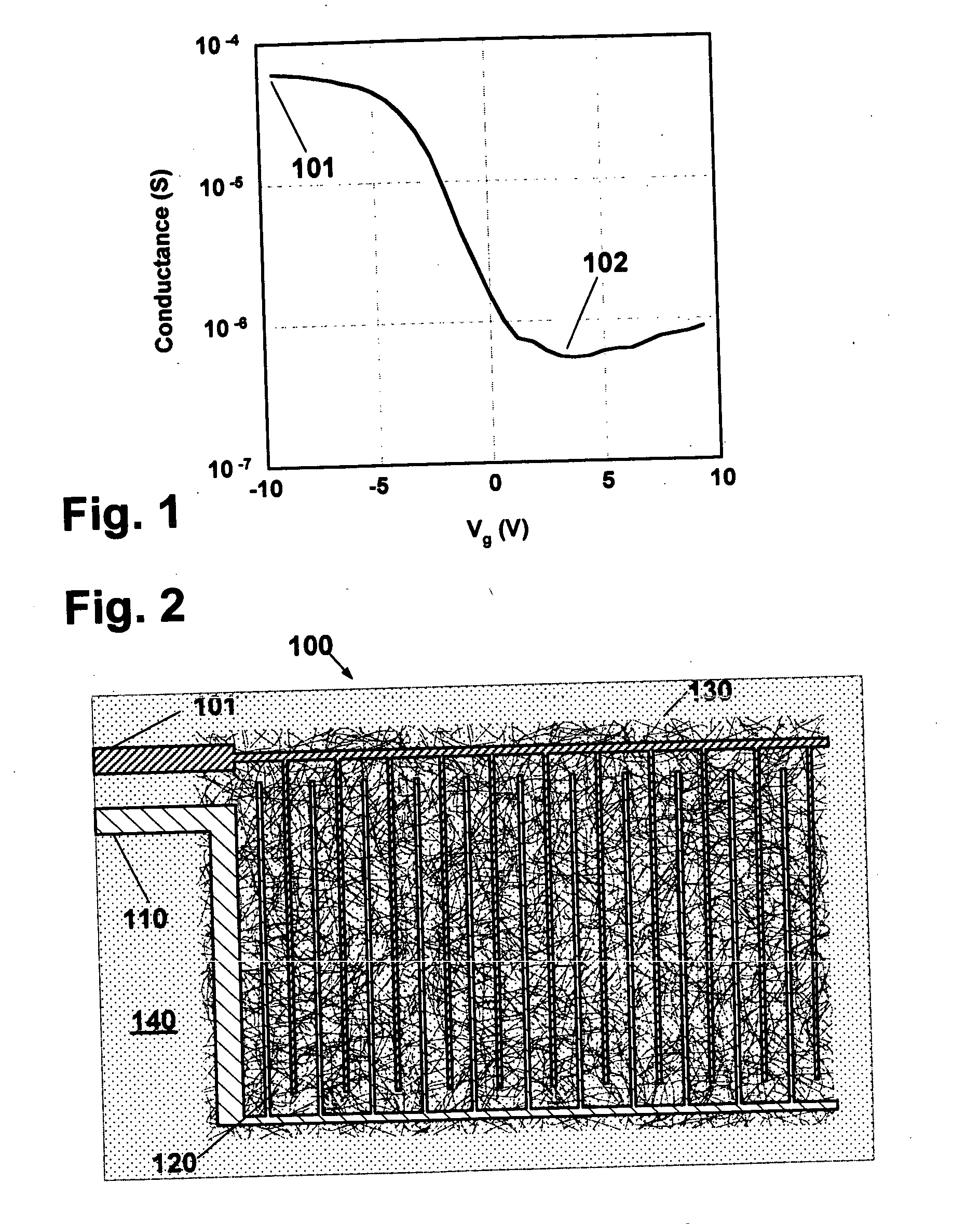
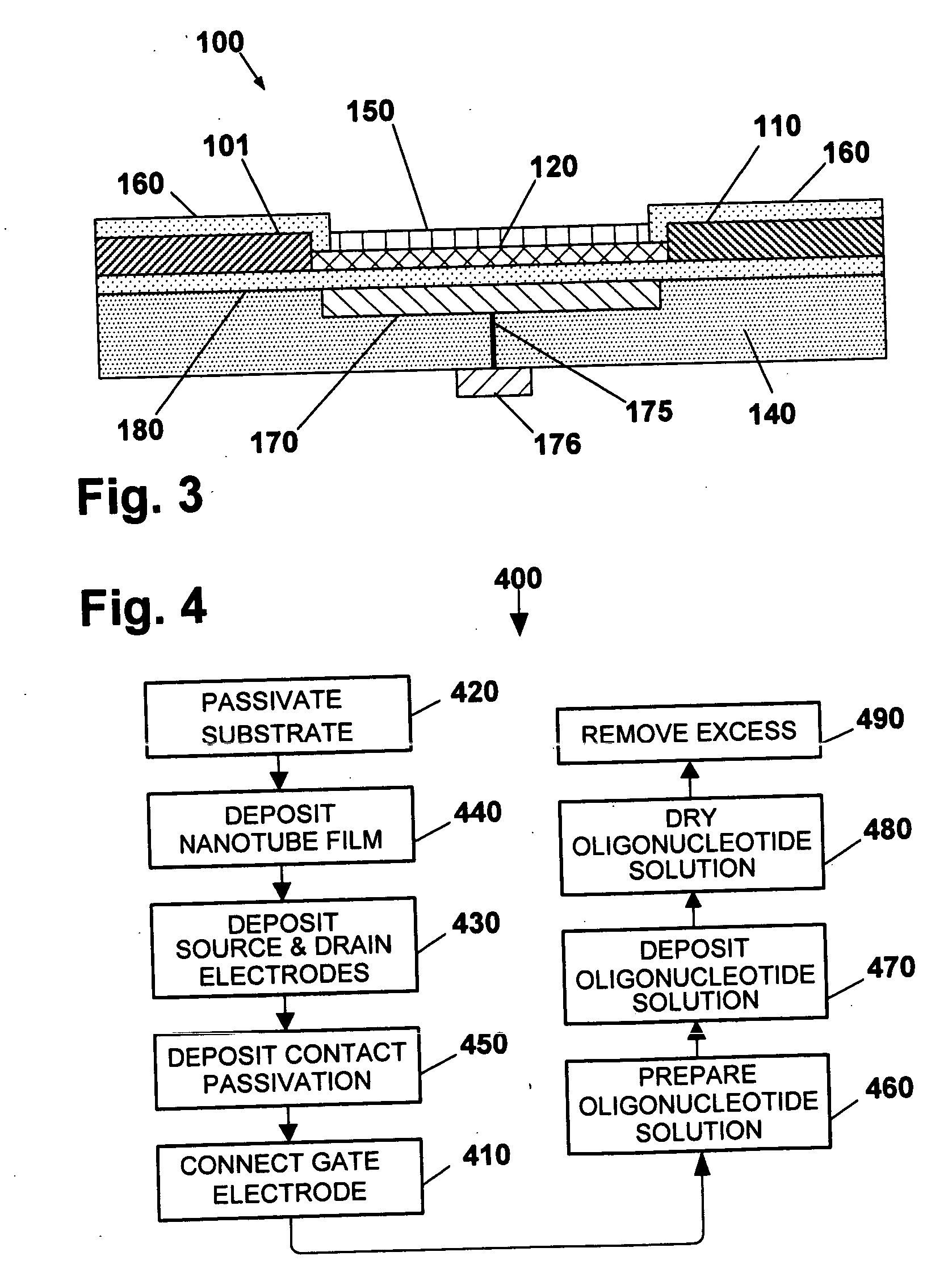
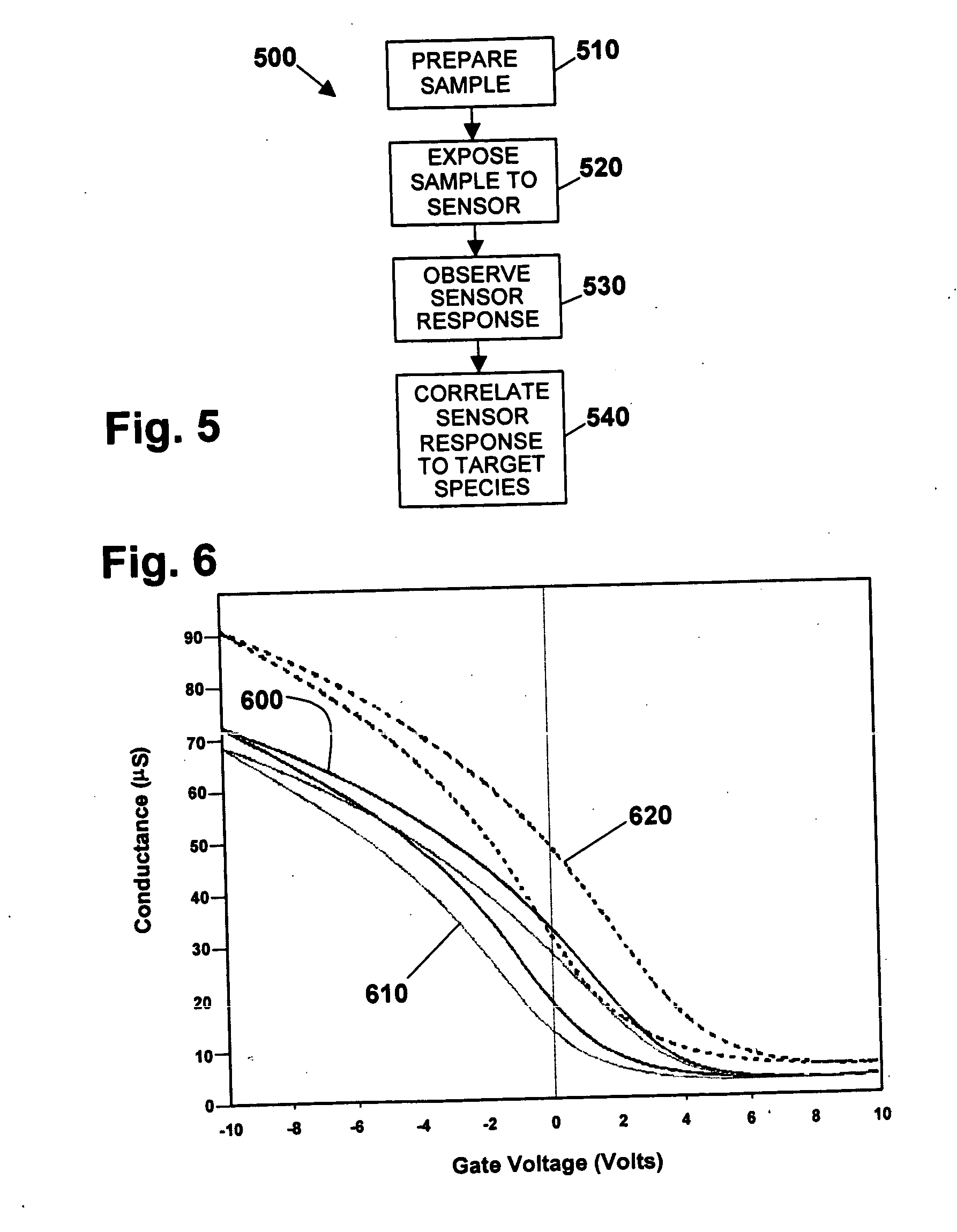
Nanotube sensor devices for DNA detection
InactiveUS20070178477A1Bioreactor/fermenter combinationsBiological substance pretreatmentsDNA SolutionsSingle strand dna
A nanotube device is configured as an electronic sensor for a target DNA sequence. A film of nanotubes is deposited over electrodes on a substrate. A solution of single-strand DNA is prepared so as to be complementary to a target DNA sequence. The DNA solution is deposited over the electrodes, dried, and removed from the substrate except in a region between the electrodes. The resulting structure includes strands of the desired DNA sequence in direct contact with nanotubes between opposing electrodes, to form a sensor that is electrically responsive to the presence of target DNA strands. Alternative assay embodiments are described which employ linker groups to attach ssDNA probes to the nanotube sensor device.
Owner:NANOMIX
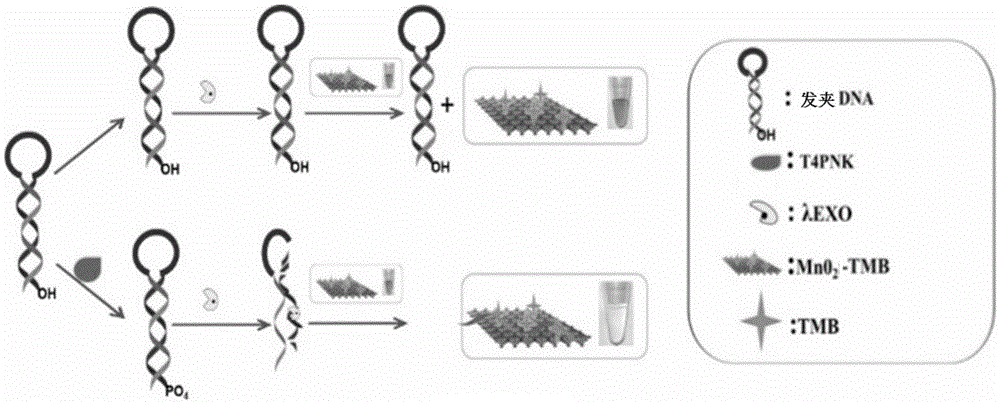
Manganese dioxide sheet mimic enzyme sensor and preparation method thereof as well as T4PNK detection method
ActiveCN105548167AHigh sensitivityLow detection limitMaterial analysis by observing effect on chemical indicatorColor/spectral properties measurementsA-DNALambda
The invention discloses a manganese dioxide sheet mimic enzyme sensor and a preparation method thereof as well as a T4PNK detection method. The preparation method comprises the following steps: (1) mixing a MnO2 nanosheet solution with a TMB (3,3',5,5'-tetramethyl benzidine) solution to obtain a MnO2-TMB mixed solution; (2) dissolving hairpin DNA in a Tris-HCl buffer solution, and adding ATP (adenosine triphosphate) and lambda EXO (lambda exonuclease) for mixing to obtain a DNA solution; and (3) mixing the MnO2-TMB mixed solution and the DNA solution to obtain the manganese dioxide sheet mimic enzyme sensor. The preparation method is simple in operation, and the sensor has the characteristics of high sensitivity, low detection limit and simple operation when used for detecting T4PNK.
Owner:ANHUI NORMAL UNIV
Analysis method for detecting diversity of soil microorganism
The invention relates to a biological analysis method, especially to an analysis method for detecting the diversity of soil microorganism. The method comprises the following steps of: (1) carrying out DNA extraction in soil; (2) detecting DNA concentration and purity in soil by using the DNA extract obtained from Step (1); (3) carrying out PCR specific amplification on the extracted DNA from soil; and (4) carrying out denaturing gradient gel electrophoresis analysis on the PCR reaction product. By the adoption of the method for detection and analysis of various soils, the result is consistent with the real state of microorganisms as there is no loss of the extracted DNA solution during the treatment process. Therefore, the invention provides a perfect solution for the research of the soil microorganism state.
Owner:SHANGHAI HAIDI GARDENING
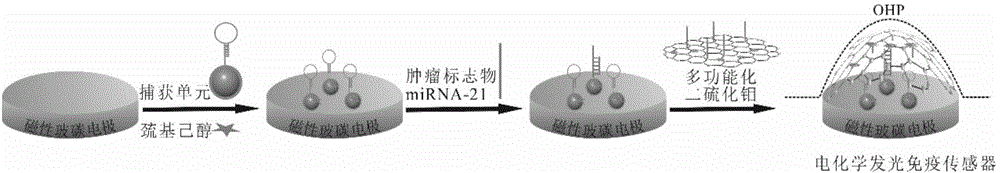
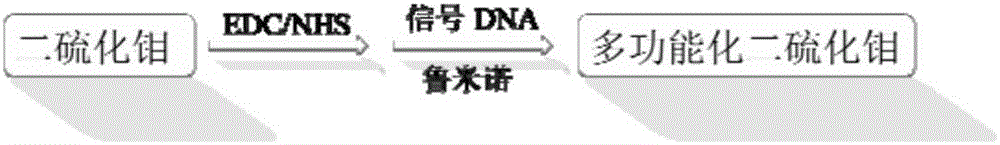
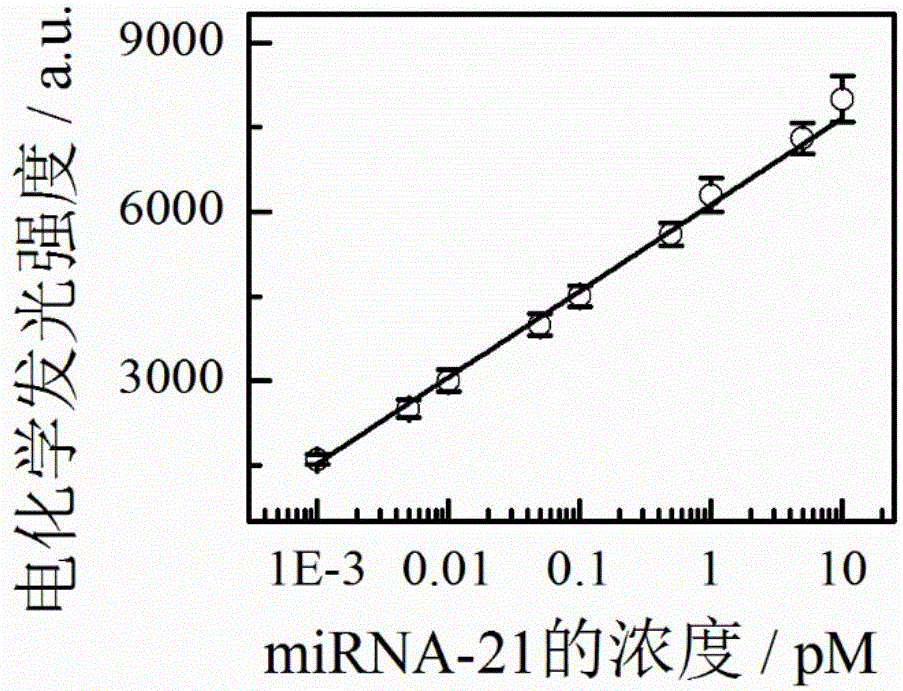
Preparation method and applications of miRNA-21 electroluminescent immunosensor based on multi-functionalized molybdenum disulfide
ActiveCN106568936AIncrease surface areaIncrease the number of markersChemiluminescene/bioluminescenceBiological testingMagnetite NanoparticlesA-DNA
The invention discloses a preparation method and applications of a miRNA-21 electroluminescent immunosensor based on multi-functionalized molybdenum disulfide. The preparation method is characterized by comprising: carrying out amination on prepared Fe3O4 magnetic nanoparticles, and reacting with chloroauric acid to prepare Fe3O4@Au magnetic nanoparticles; carrying out carboxylation on prepared monolithic layer MoS2, sequentially reacting with a coupling reagent, a signal DNA solution and a luminol solution, finally adding a mercaptohexanol solution, and carrying out vibration washing to obtain a signal unit solution; carrying out a reaction on the Fe3O4@Au and a DNA capturing solution to obtain a capture unit solution; and coating the capture unit solution on the surface of a magnetic glassy carbon electrode in a dropwise manner, and then sequentially coating miRNA-21 and the signal unit solution on the surface of the magnetic glassy carbon electrode in a dropwise manner so as to obtain the product. The miRNA-21 electroluminescent immunosensor of the present invention has advantages of high sensitivity, strong specificity, high accuracy, simple operation step, and low experimental cost.
Owner:NINGBO UNIV
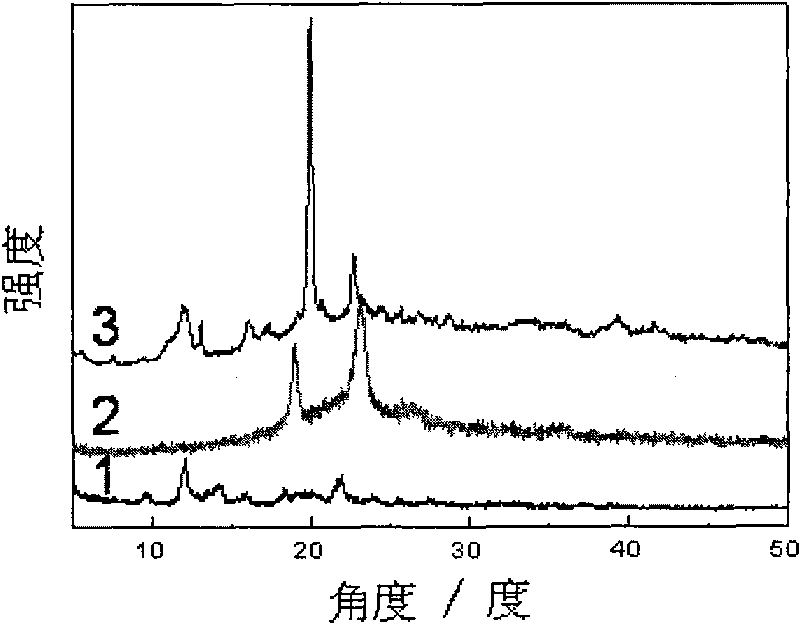
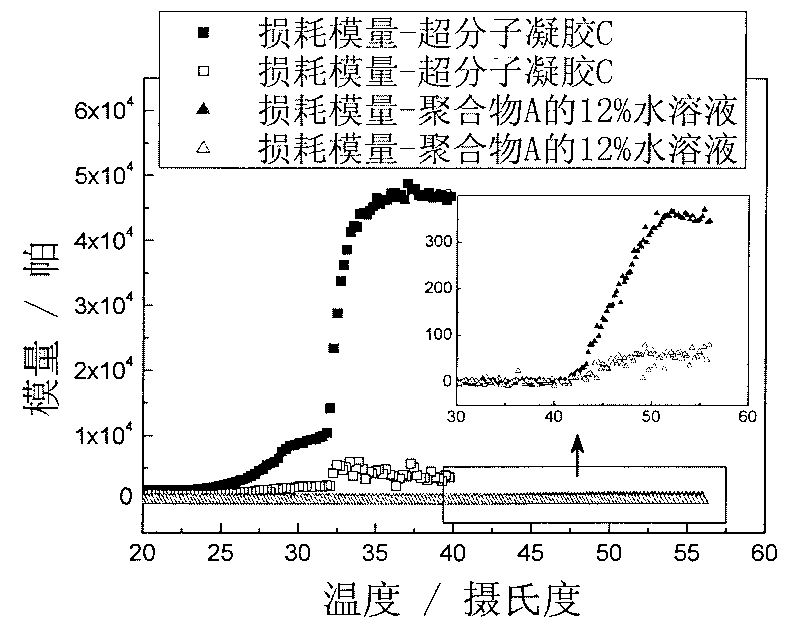
Supramolecular hydrogel gene vector material, and preparation method and application thereof
InactiveCN101716346AEasy to prepareMild conditionsGenetic material ingredientsImmunological disordersEnd-groupGene vector
The invention discloses a supramolecular structural hydrogel gene vector material and a preparation method and application thereof. The method comprises the following operation steps: synthesizing a multi-block copolymer with cation chain segments through modification of a terminal group of triblock copolymer of a polyethylene glycol-polypropylene glycol-polyethylene glycol which is modified by polylysine; then, mixing the copolymer with DNA solution to obtain DNA compound micelle; further, mixing and then stirring the micelle solution and solution of alpha-cyclodextrin; and keeping mixed solution at the room temperature to obtain hydrogel. The hydrogel can be used for preparing an injectable gene vector. The method has the advantages that the method has simple operation, can adjust the strength of the hydrogel and the gelatination time, can mold at the room temperature and does not relate to chemical crosslinking reaction and the use of organic solvent; and the obtained hydrogel has the advantages of temperature sensitivity, good bio-compatibility, obvious transfection effect and the like; and the method is hopeful to be widely applied in the field of a biomedical engineering material.
Owner:SUN YAT SEN UNIV
Fractionation of macro-molecules using asymmetric pulsed field electrophoresis
InactiveUS20050161331A1Rapid fractionationElectrostatic separatorsSludge treatmentDNA SolutionsPulsed field electrophoresis
A method and apparatus for fractionation of charged macro-molecules such as DNA is provided. DNA solution is loaded into a matrix including an array of obstacles. An alternating electric field having two different fields at different orientations is applied. The alternating electric field is asymmetric in that one field is stronger in duration or intensity than the other field, or is otherwise asymmetric. The DNA molecules are thereby fractionated according to site and are driven to a far side of the matrix where the fractionated DNA is recovered. The fractionating electric field can be used to load and recover the DNA to operate the process continuously.
Owner:THE TRUSTEES FOR PRINCETON UNIV
Carbon nano-tube film supported on an aluminum substrate and preparation method thereof
ActiveCN101532132AConductiveTranslucentAnodisationSolid/suspension decomposition chemical coatingDNA SolutionsCarbon nanotube
The invention relates to a carbon nano-tube film supported on an aluminum substrate and a preparation method thereof, wherein, the substrate is an anodized aluminum substrate with the pore diameter of the oxide film being 15nm to 200nm and the thickness thereof being 5 to 150 Mum; the film supported on the aluminum substrate comprises a carbon nano-tube with the diameter being 0.5nm to 20nm and single-stranded DNA (deoxyribonucleic acid) which is 5 to 60 guanines (G) or 5 to 60 thymines (T) or the combination of the two bases; the thickness of the carbon nano-tube film is 2nm to 500nm; and the square resistance value is 0.5k to 50k ohm / sq. The preparation method comprises the following procedures: soaking the anodized aluminum substrate into the DNA solution of a single-walled carbon nano-tube for 0.5h to 96h; and self-assembling on the surface of the substrate to form the carbon nano-tube conducting film. The invention has the advantages that the preparation is simple, and the obtained carbon nano-tube conducting film is uniform and stable with good conductivity and certain light transmittance.
Owner:TIANJIN UNIV
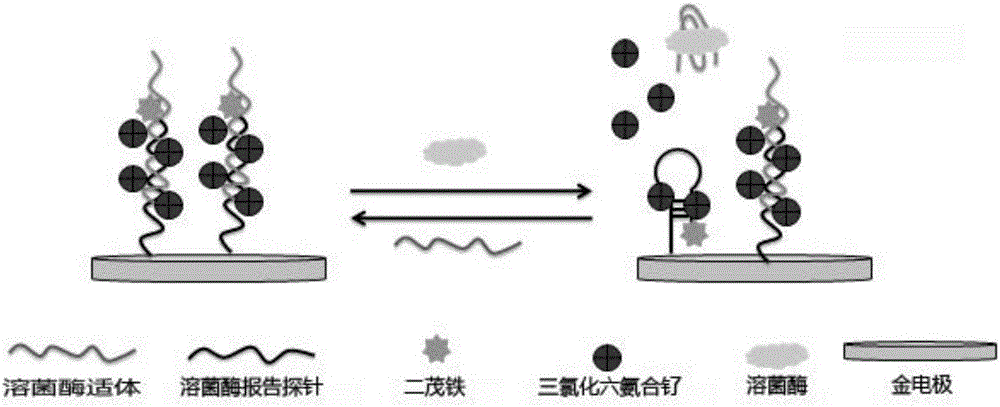
Electrochemical biosensor used for detecting lysozyme and a preparation method thereof
The invention discloses a preparation method of an electrochemical biosensor used for detecting lysozyme. The preparation method includes the following steps that step1, a reporter probe DNA chain S1 is designed; step2, a gold electrode is pretreated; step3, the disulfide bond of S1 is opened in a buffering solution, and then lysozyme aptamers S2 are added so that S2 and S1 can form double-chain DNA; step4, the treated gold electrode is soaked in the double-chain DNA solution, and the double-chain DNA is arranged on the surface of the gold electrode in a modified mode. The invention further discloses the electrochemical biosensor prepared through the method and application of the sensor in the process of detecting lysozyme. The obtained electrode is soaked in hexaammineruthenium (III) chloride detection solution, then the obtained electrode, a calomel reference electrode and a platinum wire contrast electrode form a three-electrode system, and SWV detecting is performed. For the first time, a strategy of amplifying double electrochemical signals is combined with a label-free strategy to be applied to detecting the lysozyme, and therefore the lysozyme is detected in a high-sensitivity mode.
Owner:QINGDAO UNIV
Noninvasive fetus 21, 18 and 13 trisomic syndrome antenatal detection positive quality control product and preparing method thereof
InactiveCN104531842AEasy to storePrevent deviationMicrobiological testing/measurementTrisomy 13 SyndromeTrisomy X syndrome
The invention provides a noninvasive fetus 21, 18 and 13 trisomic syndrome antenatal detection positive quality control product and a preparing method thereof, and belongs to the fetus chromosome aneuploid noninvasive antenatal detection range. The preparing method of the positive quality control product includes the steps that DNAs of abortion groups of fetuses suffering from 21, 18 and 13 trisomic syndromes are broken into DNA fragments with the length ranging 160 bp to 200 bp; the abortion group DNA fragments recycled after breaking are added into mixed plasma in proportion; and the total plasma free DNAs are finally extracted, and the positive quality control products with different concentrations are prepared after attenuation with ultrapure water is carried out. The positive quality control product is a white transparent DNA solution, and has the beneficial effects of being stable, easy to store, convenient and rapid to use, non-toxic and the like; in addition, deviations of detection results of other samples in the same batch can be effectively avoided; and the accuracy and the reliability of the 21, 18 and 13 trisomic syndrome noninvasive antenatal detection are improved.
Owner:DAAN GENE CO LTD
Method for carrying out nucleic acid isothermal amplification by using paper-based microfluid
ActiveCN104293945AReduce weightEasy to carryMicrobiological testing/measurementDNA SolutionsReaction layer
The invention discloses a method for carrying out nucleic acid isothermal amplification by using paper-based microfluid. The method comprises the following steps: (1) preparing a sample DNA solution; (2) coating a primer on a reaction zone of paper-based microfluid, wherein the microfluid comprises a reaction layer made of filter paper and at least one sampling layer; sampling holes are formed at the centers of the sampling layers; a plurality of reaction holes are evenly distributed around the sampling holes; each orifice is coated by a hydrophobic material; the sampling layers are stacked above the reaction layer; the reaction layer is provided with a sample zone, a plurality of reaction zones and reaction channels; the sample zone is drawn by the hydrophobic material and corresponds to the sampling hole; the plurality of reaction zones and reaction channels correspond to the reaction holes; the reaction zones are the same as the reaction holes in diameter; the diameter of the sample zone is smaller than those of the sampling holes; and the reaction zones are communicated with the sample zones through the reaction channels of which the widths are gradually reduced from the reaction zones to the sample zones; (3) dropwise adding the reaction liquid into the sampling holes, heating the microfluid to reach amplification temperature; and (4) observing the color change of the reaction zones, so as to obtain the detection result. The method is low in cost and accurate in result.
Owner:THE FIRST AFFILIATED HOSPITAL OF THIRD MILITARY MEDICAL UNIVERSITY OF PLA
Method for quickly extracting DNA (deoxyribonucleic acid)
The invention relates to the technical field of biology, in particular to a method for quickly extracting DNA (deoxyribonucleic acid). The method for extracting DNA is chip and easy to operate and comprises the following steps: a, sequentially adding protein precipitate reagent, pyrolytic reagent and a biological sample containing DNA into a container, and uniformly mixing to release cell contents, and agglomerating proteins to obtain lysate which contains the DNA component and non-DNA component released from the biological sample, wherein the protein precipitate reagent is polyvalent metal salt; and b, separating the DNA and non-DNA components in the lysate obtained in the step a to obtain the purified DNA solution. The method has the advantages: protein contamination can be removed by adopting a high-efficiency protein precipitate reagent, the cell lysis and the protein precipitate are synchronously performed. Compared with the prior art, the method has simpler steps and high efficiency, and the use cost is further reduced.
Owner:亚能生物技术(深圳)有限公司
Method for extracting soil microbial DNA
InactiveCN1978453ALow humusReduce organic acid contentSugar derivativesSugar derivatives preparationDNA SolutionsMicroorganism
This invention relates to an extracting method of soil microorganism DNA. The stated method includes steps as following: cell disruption of soil microorganism, obtaining crude DNA solution, obtaining soil microorganism DNA; the stated method of this invention is using PVPP, protease K etc for valid combination. After organic solvent such as PEG20000, isopropyl alcohol and so on are precipitated, use spectrophotometer to determine value of OD260 / OD230, OD260 / OD280 close to standard value. It can be directly used for molecule operation, possessing great application perspective.
Owner:ZHEJIANG UNIV OF TECH
Method for fixing DNA basic group to surface of glass slide
ActiveCN104142262AImprove wettabilityIncrease fix ratePreparing sample for investigationDNA SolutionsAlcohol
The invention discloses a method for fixing a DNA basic group to the surface of a glass slide. The method is suitable for fixing the DNA basic group to the surface of the glass slide. Firstly, the glass slide is cut into small pieces, washed, immersed in 95 percent of HCL and 99.7 percent of absolute ethyl alcohol mixed solution, processed through acid, and then is washed. Secondly, the glass slide is immersed in an APTES absolute ethyl alcohol to be prepared into a silanized glass slide. Thirdly, the silanized glass slide is immersed into a glutaraldehyde glutaraldehyde water solution to be reacted and flushed and is aired to be used later. Fourthly, a buffer solution is prepared, cosolvent and a surface active agent are added to the buffer solution to prepare a surface active agent solution, the DNA basic group is added to the surface active agent solution, full stirring is carried out to enable the DNA basic group to dissolve completely, and a DNA solution is prepared. Fifthly, the DNA solution is sampled to be dripped on the glass slide, the sampled DNA solution is cultivated in a calorstat at the appropriate temperature and is cleaned, and fixing of the DNA basic group is finished. By means of double modification of the glass slide, the wettability of the glass slide is improved, and the fixing rate of the DNA basic group is improved. Representation is carried out on the DNA basic group through a laser confocal raman spectrometer, the steps, needed in light splitting method of fluorescence, of fluorescence labeling and gene amplifying and hybridizing can be removed, and the DNA basic group fixing steps are greatly simplified.
Owner:CHINA UNIV OF MINING & TECH
Three-branch-like nucleic acid nano silver fluorescent cluster as well as preparation method and application
InactiveCN108359448AGood fluorescent effectHigh sensitivityMaterial nanotechnologyFluorescence/phosphorescenceDNA SolutionsPhosphate
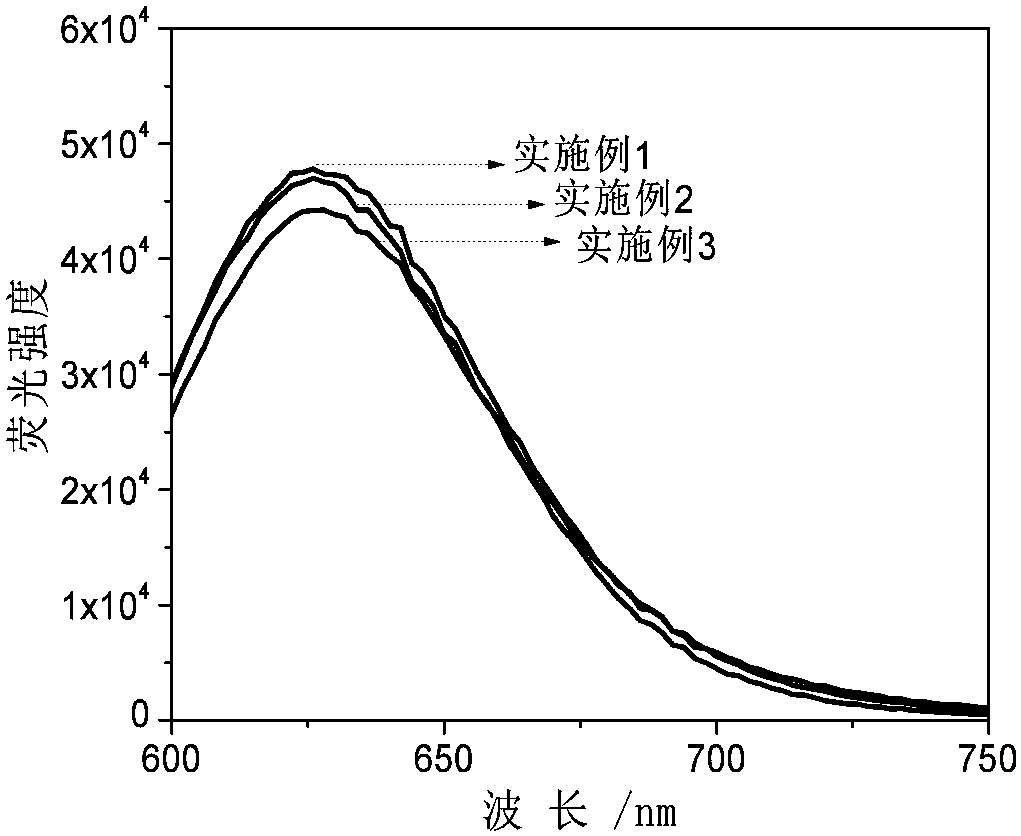
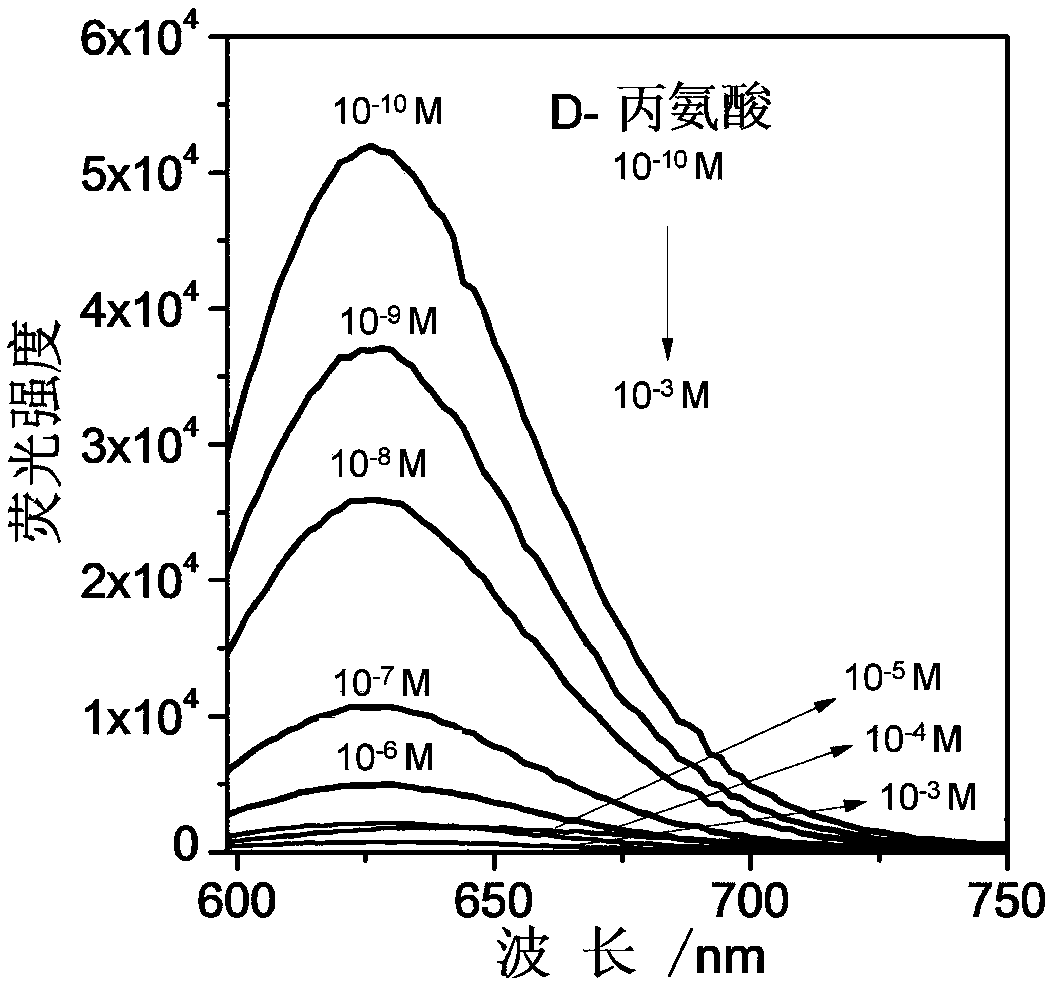
The invention discloses a three-branch-like nucleic acid nano silver fluorescent cluster as well as a preparation method and an application. The preparation method comprises the steps as follows: (1)three oligonucleotides are designed and synthesized according to a base complementary pairing principle to be used for forming three-branch-like DNA with the same sticky ends on all branches; (2) thethree oligonucleotides are prepared into corresponding aqueous solutions; (3) a three-branch-like DNA solution with the same sticky ends on all branches is prepared; (4) the three-branch-like DNA solution with the same sticky ends on all branches, a silver nitrate aqueous solution, a phosphate buffered solution and a sodium borohydride aqueous solution are stirred, and the three-branch-like nucleic acid nano silver fluorescent cluster is obtained. The three-branch-like nucleic acid nano silver fluorescent cluster is stronger in fluorescent effect and stable, the sensitivity of a fluorescent material in analysis and detection is improved, and the application range of the nucleic acid nano silver fluorescent material is expanded. The nucleic acid nano silver fluorescent cluster can detect Dtype amino acid.
Owner:TIANJIN UNIV
Simple sequence repeat-polymerase chain reaction (SSR-PCR)-based hybrid rape seed purity detection method
InactiveCN102321767AGood polymorphismStrong specificityMicrobiological testing/measurementDNA SolutionsBuffer solution
The invention discloses a simple sequence repeat-polymerase chain reaction (SSR-PCR)-based hybrid rape seed purity identification method, which comprises the following steps of: performing sprouting culture on hybrid rape sample seeds to be detected, performing alkaline lysis on the cultured seedlings, simultaneously performing ultrasonic disruption treatment, and adding an extracting buffer solution to obtain a genome DNA solution; performing PCR amplification on genome DNA by using an SSR primer sequence; performing voltage stabilizing electrophoretic separation on a PCR amplification product in agarose gel; performing imaging and tape reading on the PCR amplification product subjected to electrophoretic separation in a gel imaging system, comparing band characteristics of the sample seeds with those of parent seeds, counting seeds with the band characteristics of male parent and the band characteristics of female parent in the sample seeds, and obtaining the purity of the hybrid rape seeds to be detected according to a variety purity formula. The identification method has the advantages of quickness, simplicity, convenience, high throughput, low detection cost, high detection efficiency, stable and reliable detection results and the like.
Owner:湖南省作物研究所
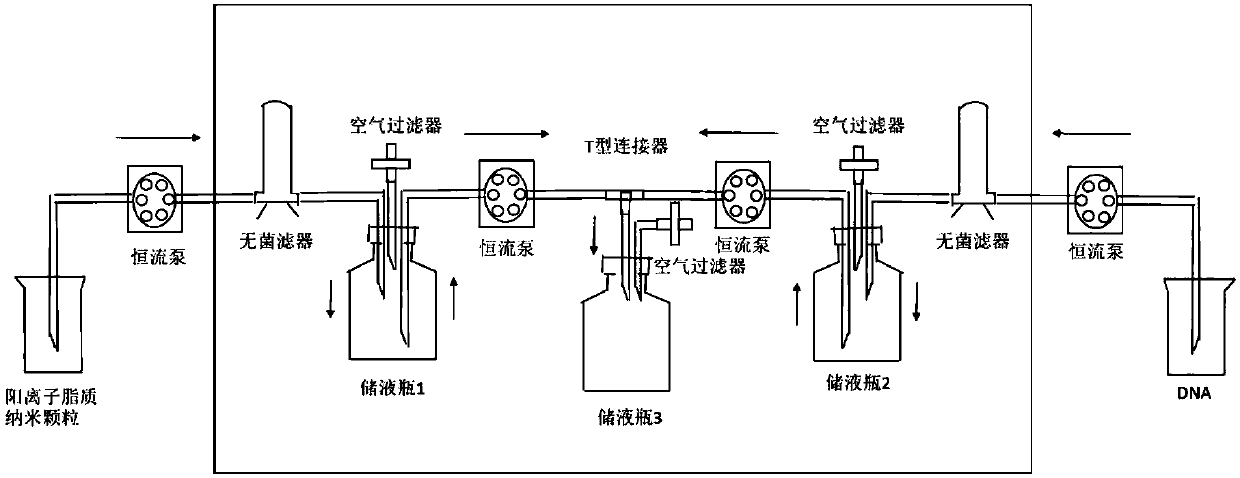
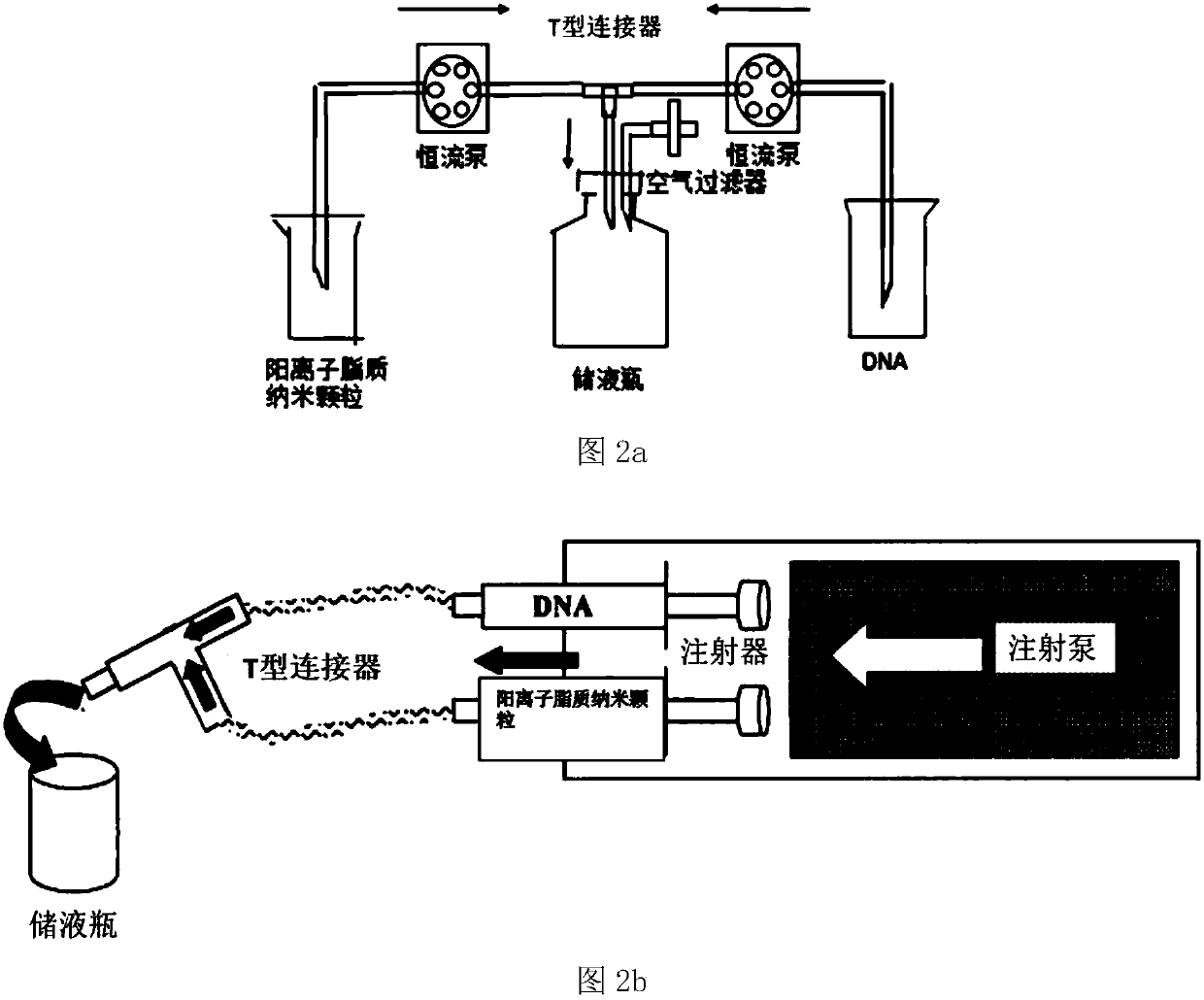
Positive ion lipid nanometer particle/DNA compound and preparation method thereof
ActiveCN109893664AEvenly distributedMonodisperseGenetic material ingredientsPharmaceutical non-active ingredientsDNA SolutionsAlcohol
The invention provides a positive ion lipid nanometer particle / DNA compound and a preparation technology thereof. The preparation technology comprises the following steps that (1) a positive ion lipidmaterial is dissolved in absolute ethyl alcohol through heating; (2) an ethyl alcohol solution prepared in the step (1) is added to an aqueous phase solution dropwise, and positive ion lipid nanometer particles are formed through self-assembling; (3) residue ethyl alcohol in the positive ion lipid nanometer particles of the step (2) is removed; (4) filtering is carried out; (5) a DNA solution isprepared; (6) the prepared positive ion lipid nanometer particles in the step (4) and the DNA solution prepared in the step (5) are mixed according to a certain weight ratio to form the positive ion lipid nanometer particle / DNA compound; and (7) filtering is carried out. According to the preparation technology of the positive ion lipid nanometer particle / DNA compound, the operation of the preparation method is easy and rapid, the particle diameter of the prepared positive ion lipid nanometer particle / DNA compound is 50-150 nm, PDI<0.3, the positive ion lipid nanometer particle / DNA compound isdistributed in a mono-dispersion mode, the structure is stable, the compound is subjected to filtering sterilization at a terminal, and the security of the compound in clinic application of preparation of tumor curing medicine is effectively ensured.
Owner:SICHUAN UNIV
Method for detecting cocaine by using quartz crystal microbalance
InactiveCN102967523AWith real-time detectionSuitable for large-scale applicationsWeighing by absorbing componentDNA SolutionsQuartz crystal microbalance
Owner:SUZHOU INST OF NANO TECH & NANO BIONICS CHINESE ACEDEMY OF SCI
Method for rapid detection of listeria monocytogenes with high sensitivity and kit thereof
ActiveCN102321762AShorten the timeHigh sensitivityMaterial nanotechnologyMicrobiological testing/measurementDNA SolutionsBiological cell
The invention discloses a method for rapid detection of listeria monocytogenes with high sensitivity and a kit thereof, and the method comprises the following steps: (1) lysing biological cells to release DNA; (2) mixing the solution containing the biological DNA and magnetic nano-particles which can be bonded with the DNA, wherein the magnetic nano-particles are gamma-Fe2O3 nano-particles coated by carboxyl-modified silica, allowing the DNA to be bonded onto the surfaces of the magnetic nano-particles under the action of isopropanol, collecting the magnetic nano-particles under the action of an external magnetic field to obtain DNA-bonded magnetic nano-particles; (3) adding water, performing magnetic separation, collecting the aqueous solution to obtain the separated biological DNA solution; (4) performing PCR amplification by a specific primer of a target organism with the DNA solution as a template. The whole detection process of the invention only need 2 hours; the detection limit of listeria monocytogenes is about 1 CFU / 25 g (mL); the time required is short; the sensitivity is high; and the results are reliable.
Owner:上海德诺产品检测有限公司
Optical means for detecting i-motif conformation of DNA
ActiveCN101434984ASimplify detection stepsShorten detection timeMicrobiological testing/measurementColor/spectral properties measurementsDNA SolutionsTelomeric dna
The invention discloses an optical method for detecting the four-chain-motif conformation of DNA, comprising the following steps: 1) the cationic water-soluble conjugated polymer solution with equivalent charge amount is added to DNA solution; 2) and then colorimetric and / or fluorescence detection are / is carried out to the DNA solution, and finally the C-rich sequence conformation of the DNA to be detected, such as human telomere DNA, can be judged according to the color change of the solution, fluorescence spectra or UV-visible absorption spectrum of the solution. The invention has simple method and high selectivity and sensitivity, and has important significance in biological detection, early diagnosis and treatment of diseases.
Owner:SHANGHAI INST OF APPLIED PHYSICS - CHINESE ACAD OF SCI
Real-time fluorescent RCR molecular detection kit for leptosphaeria maculans and detection method thereof
InactiveCN101928779AQuick checkSensitive detectionMicrobiological testing/measurementPositive controlFluorescence
The invention relates to a real-time fluorescent RCR molecular detection kit for leptosphaeria maculans, which contains a general hybrid reagent for gene expression, ddH2O, a probe and primer mixture and a positive control. The probe and primer mixture contains 5mumol / L of LMpro probe, 18mumol / L of LMf upstream primer and 18mumol / L of LMr downstream primer; and the positive control is 100ng / muL leptosphaeria maculans DNA solution. The kit can directly detect the sick rape seed or plant residues, has the advantages of quickness, sensitivity, accuracy and good repeatability, particularly can be used for detecting a sample with low pathogenic bacteria content, is particularly suitable for quickly detecting the leptosphaeria maculans in seeds, plants and sick residues of cruciferous crops such as imported and exported rape seeds and the like, and has good application prospect.
Owner:ANIMAL AND PLANT & FOOD DETECTION CENTER JIANGSU ENTRY EXIT INSPECTION AND QUARANTINE BUREAU
DNA sensitivity electrode modified by hydrotalcite nanometer slice and preparation thereof
InactiveCN101281158AWide linear response rangeHigh detection sensitivityMaterial analysis by electric/magnetic meansModified dnaElectrochemical biosensor
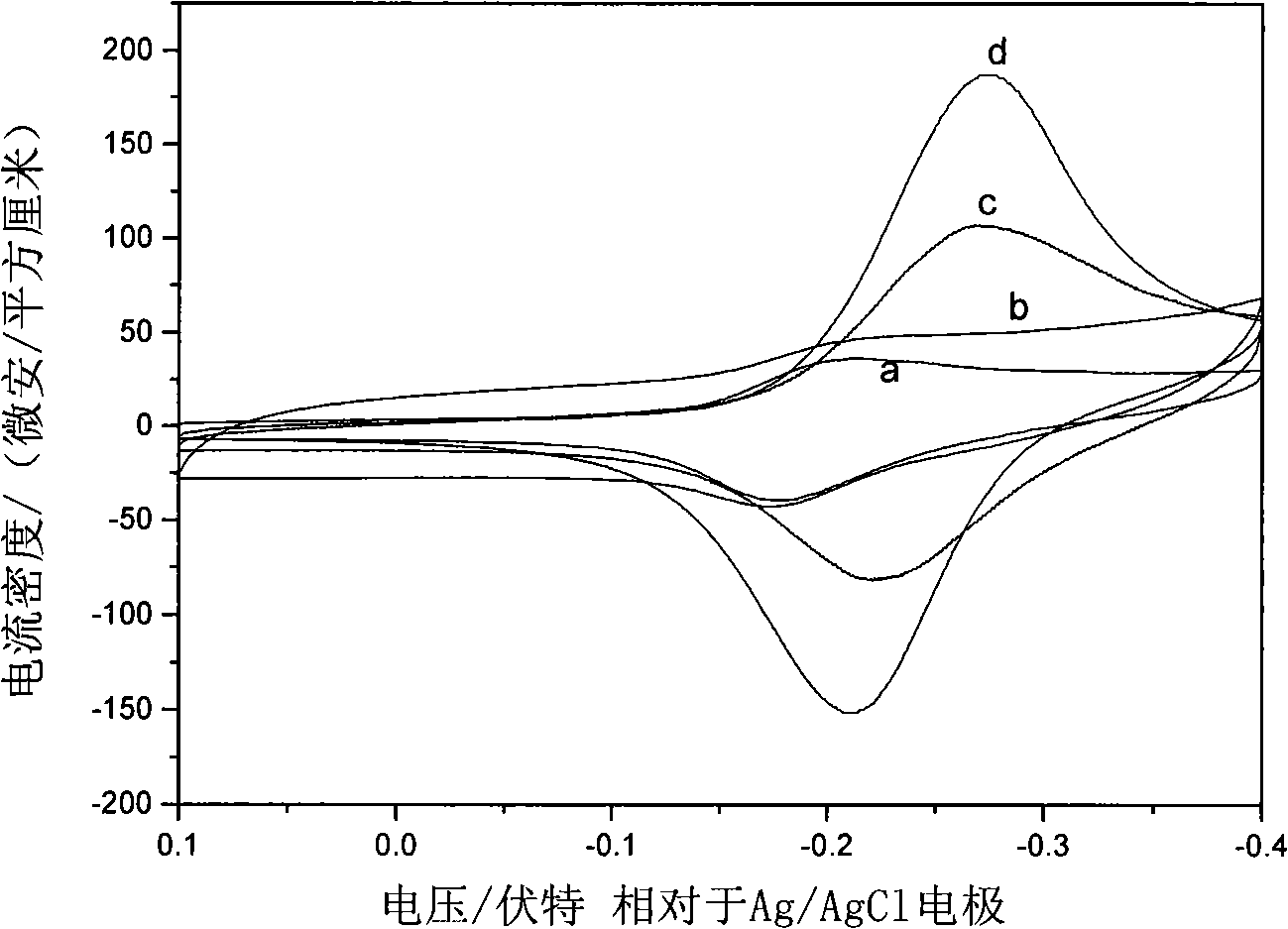
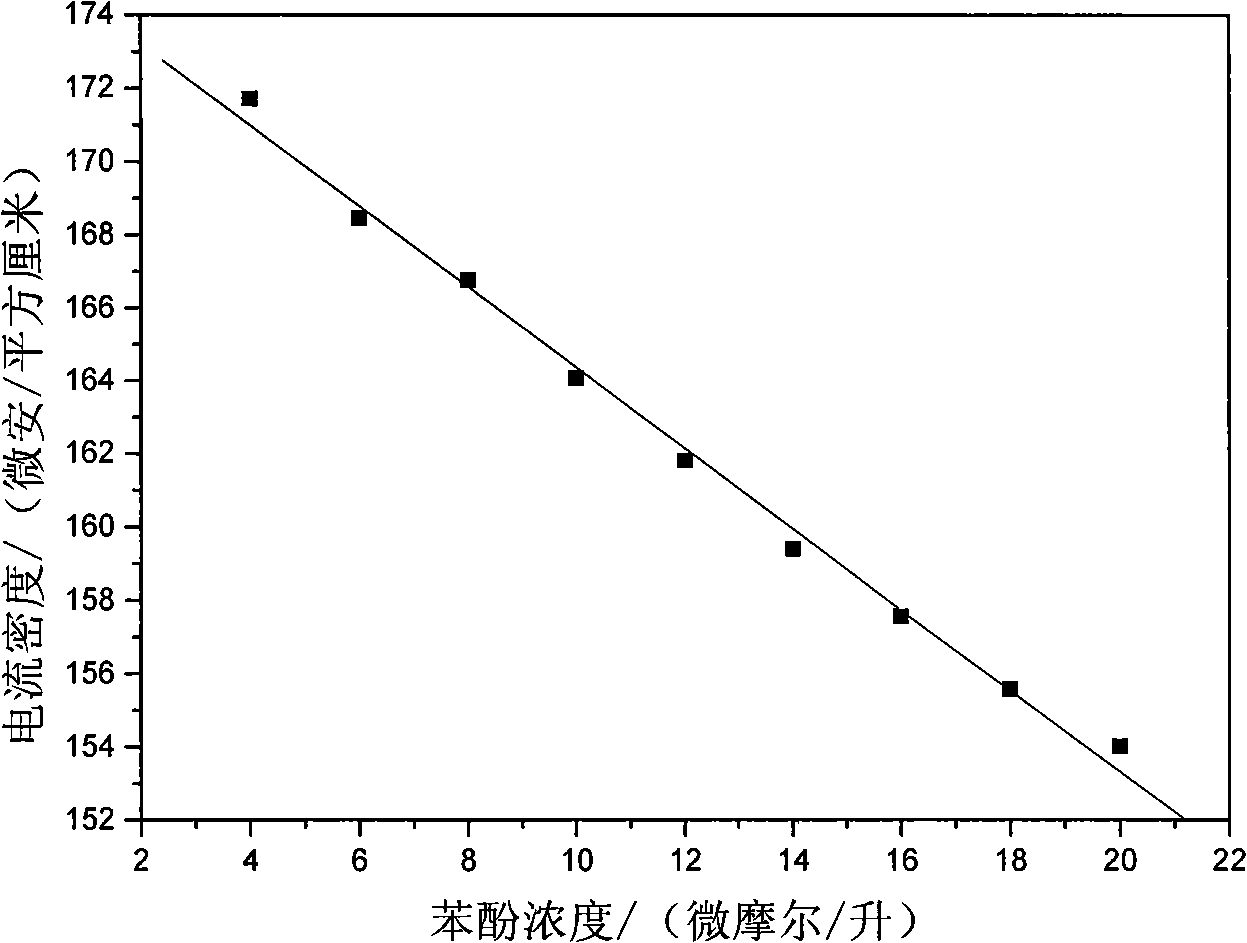
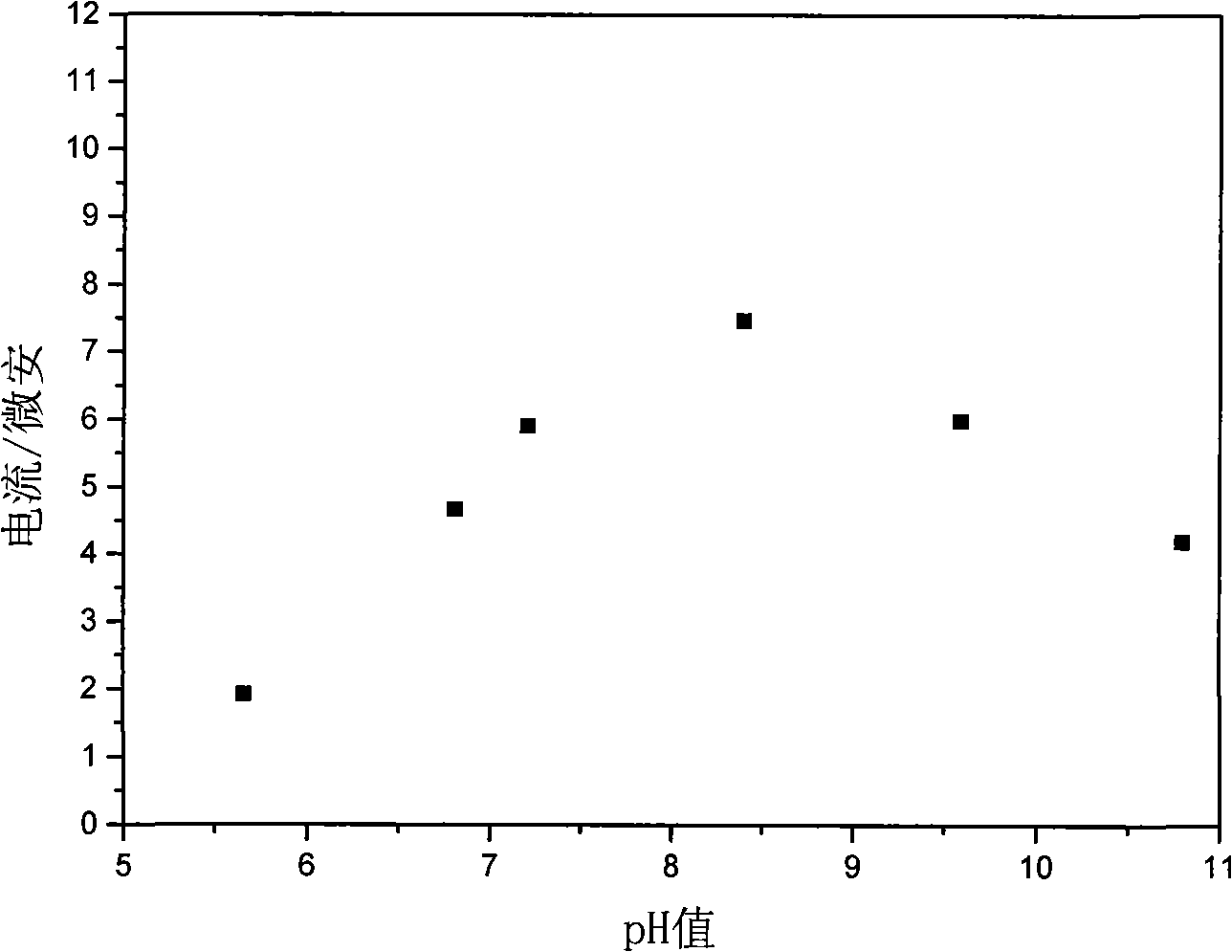
The invention relates to a DNA sensitive electrode decorated by hydrotalcite nano-chip and the preparation method thereof, which belongs to the field of electrochemical biosensors and preparation technology thereof. The sensitive electrode uses a glassy carbon electrode as the substrate electrode, and a sensitive film composed of double-strand DNA and co-aluminum hydrotalcite nano-chip is decorated on the surface of the substrate electrode. The preparation method of the sensitive electrode is as follows: the co-aluminum hydrotalcite nano-chip collosol is coated on the surface of the clean glassy carbon electrode; after dried at room temperature, the glassy carbon electrode is washed with second distilled water and dried, then the double-strand DNA solution is dropped on the surface of the hydrotalcite nano-chip; after dried at room temperature, the glassy carbon electrode is washed with second distilled water and dried, then the DNA sensitive electrode decorated by hydrotalcite nano-chip is prepared. The sensitive electrode has the advantages that the invention uses the properties of the hydrotalcite nano-chip, that is, the hydrotalcite nano-chip has large surface area, has high-density positive charge, has a lot of hydroxide radical on the surface and has good electric conductibility and so on, to realize the stability and fixing of mass DNA, and the electrode can be used in quantitative determination of phenol, which has good anti-interference performance.
Owner:BEIJING UNIV OF CHEM TECH
Detection kit and detection method for Enteromorpha compressa
InactiveCN102051417AThe result is accurateAccurate separationMicrobiological testing/measurementForward primerDNA Solutions
The invention discloses a detection kit and detection method for Enteromorpha compressa. The detection kit for Enteromorpha compressa comprises 10 PCR (polymerase chain reaction) buffer solutions, an MgCl2 solution, a dNTP (deoxynucleotide triphosphate) solution, a Taq DNA (deoxyribonucleic acid) polymerase solution, a forward primer and a reverse primer in the volume ratio of 2.5:1.5:2:0.2:1:1, wherein the nucleotide sequence of the forward primer is 5'-CGTTTTCGGA ACCGCCGGTGA-3', and the nucleotide sequence of the reverse primer is 5'-GGCCAGGTCCACGGCCCGCTCT-3'. The detection method for Enteromorpha compressa comprises the following steps: extracting DNA to obtain a template DNA solution, wherein the template DNA solution and the solutions in the detection kit constitute a PCR system; carrying out PCR to obtain a PCR product, and carrying out agarose gel electrophoresis on the PCR product; and quickly and simply determining whether the sample is Enteromorpha compressa according to the fact whether a 330bp specific segment strip appears, and accurately separating the Enteromorpha compressa from other seaweed samples. The invention can be used for detecting Enteromorpha compressa mature algae with multiple distinguishing features, and can also be used for quickly distinguishing seedlings with fewer morphological features and Enteromorpha compressa algae with other growth morphologies.
Owner:INSPECTION & QUARANTINE TECH CENT OF NINGBO ENTRY EXIT INSPECTION & QUARANTINE BUREAU +1
Method for extraction and purification of microbial total DNA of compost and buffer solution thereof
InactiveCN101550413AReduce pollutionIdeal anti-corrosion effectSugar derivativesDNA preparationMicroorganismDNA Solutions
The invention discloses a method for the extraction and the purification of microbial total DNA of a compost and a buffer solution thereof, the method comprises the following steps of: adding a dehumification solution in an amount of 8-15ml / g of the compost into a sample of the compost in which the DNA is to be extracted, for dehumification; then adding a DNA extraction buffer solution in an amount of 1.0-1.5ml / g of the compost for the crude extraction of the microbial total DNA; and finally, purifying the crudely-extracted DNA solution with a kit so as to obtain the microbial total DNA of the compost. The inventive method can reduce the influence of humic acids and effectively improve the yield and the purity of the DNA extracted from the compost, thus better facilitating compost microbial molecular ecology and researches of other functional genes.
Owner:HUNAN UNIV
Extraction method of strawberry genome DNA
InactiveCN104805071AQuality improvementGood amplification resultDNA preparationSodium acetateWater baths
The invention belongs to the technical field of molecular biology, and discloses an extraction method of strawberry genome DNA. The extraction method comprises following steps: liquid nitrogen is used for smashing samples into powder; CTAB extracting solution is added; an obtained mixture is subjected to water bath at 55 to 65 DEG C and then is cooled to room temperature; potassium acetate is added, and ice bath is carried out; a chloroform / isoamyl alcohol mixed liquid of a same volume is added; an obtained mixed material is mixed uniformly, and is subjected to centrifugation; the chloroform / isoamyl alcohol mixed liquid of a same volume is added into an obtained supernate A; an obtained product is mixed uniformly, is allowed to stand, and is subjected to centrifugation so as to obtain a supernate B; isopropyl alcohol is added into the supernate B, an obtained mixed solution is subjected to centrifugation so as to obtain a precipitate; a supernate C obtained via centrifugation is removed; TE buffer containing RNase is subjected to water bath at 37 DEG C; the chloroform / isoamyl alcohol mixed liquid of a same volume is added, an obtained mixed liquid is subjected to uniform mixing, is allowed to stand, and is subjected to centrifugation so as to obtain a supernate D; sodium acetate and absolute ethyl alcohol are added into the supernate D; an obtained mixed product is subjected to centrifugation so as to remove an obtained supernate E; TE buffer is added into an obtain precipitate product so as to dissolve the precipitate product, and a strawberry genome DNA solution is obtained. According to the extraction method, characteristics of CTAB and SDS extraction methods are combined, and high quality genome DNA of strawberry which is abundant in glucose and polyphenols is obtained.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
Preparing process and application of nano chitosan particle
InactiveCN1986608AGood biocompatibilityCheap sourcePharmaceutical non-active ingredientsWater bathsNon toxicity
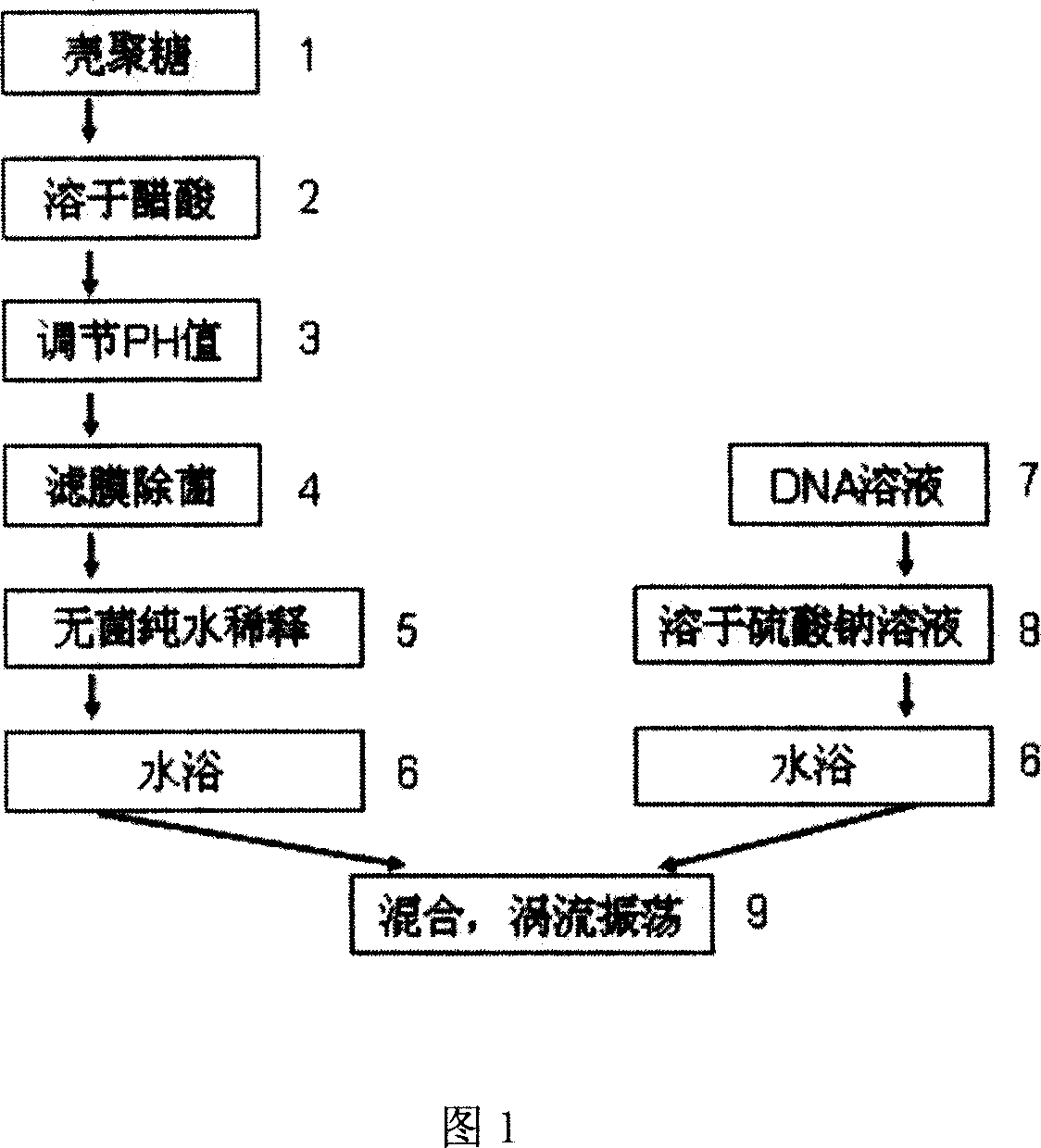
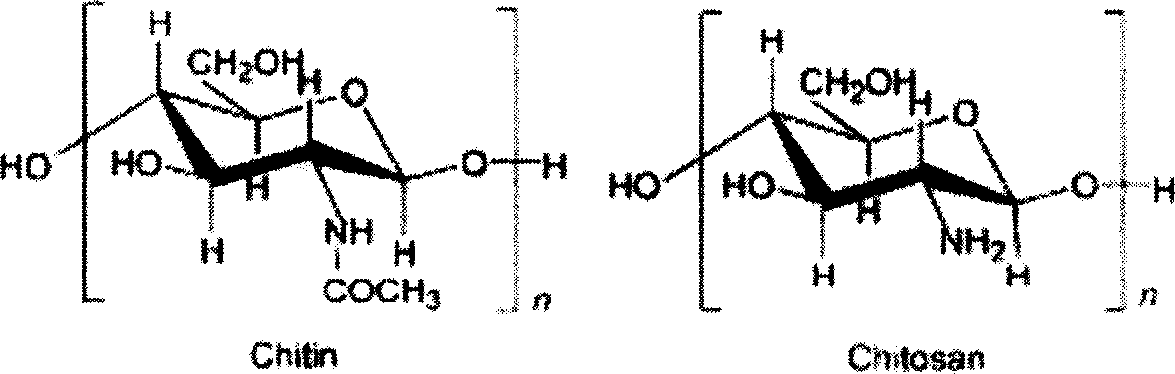
The present invention discloses preparation process and application of nanometer chitosan particle as one kind of non-viral gene transferring carrier. The preparation process is the following steps: 1. treating chitosan into chitosan solution through dissolving in acetic acid, regulating pH value, filtering membrane sterilizing and diluting with abacterial pure water; 2. treating DNA solution through dissolving 50-150 mg / l concentration DNA solution in 100 microliter in sodium sulfate solution; and 3. mixing the DNA solution and the chitosan solution in water bath through eddy flow vibration, letting stand at room temperature for 2 hr, packing and freeze drying. The nanometer chitosan particle may be used to carry several kinds of target gene for genetic treatment of diabetes. The present invention has high biocompatibility, non-toxicity, low cost and other advantages.
Owner:WUHAN UNIV
Preparation method for flaky silver nanometer materials
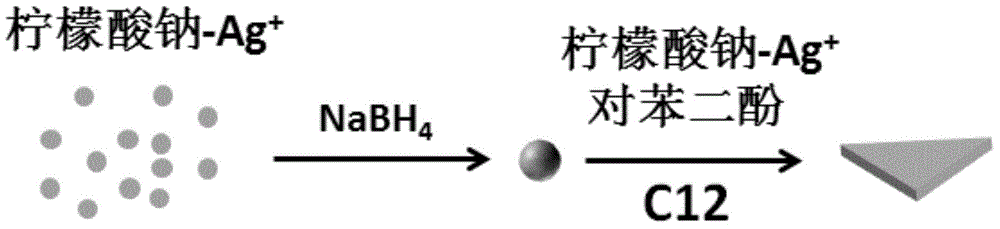
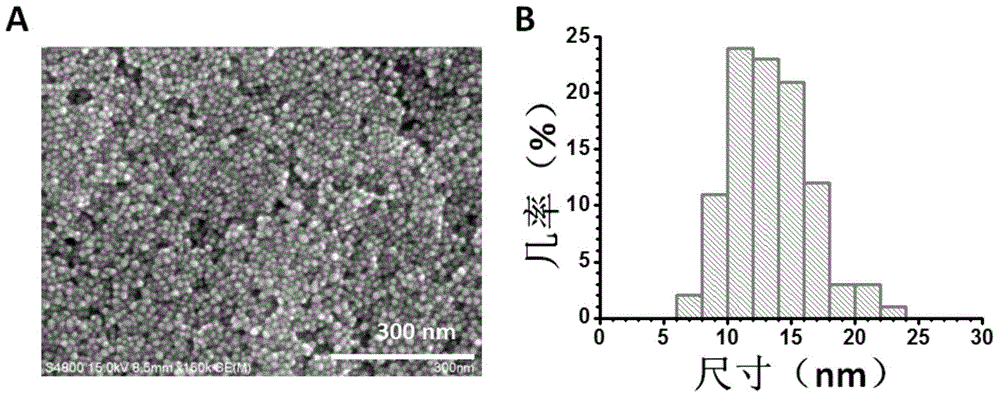
ActiveCN105149613AGood biocompatibilityReduce usageNanotechnologyDNA SolutionsBiocompatibility Testing
The invention discloses a preparation method for flaky silver nanometer materials. The method includes the following steps that firstly, at the existence of sodium citrate, a sodium borohydride solution prepared through ice water is used for reducing silver nitrate so that a silver seed solution is formed; secondly, a p-dihydroxybenzene solution, a sodium citrate buffer solution, the silver seed solution, a DNA solution, water and a silver nitrate solution are measured and taken according to the volume ratio of 3-5:3-5:1-3:1-3:27-30:9-12; and thirdly, after the p-dihydroxybenzene solution, the sodium citrate buffer solution, the silver seed solution, the DNA solution and the water are fully mixed, the silver nitrate solution is added, the mixture is kept away from light and grows for 12-14 h at the temperature of 20-24 DEG C, and then the flaky silver nanometer materials can be obtained. Compared with a traditional method, the preparation method is good in biocompatibility and high in universality, and a new platform is provided for application of the flaky silver nanometer materials in biological analysis and biomedicine.
Owner:XIAMEN UNIV
DHA-containing watermelon juice nutritious milk and preparation method thereof
InactiveCN101589744AImprove scalabilityAvoid looseMilk preparationFood preparationNutritive valuesDNA Solutions
The invention relates to a DHA-containing watermelon juice nutritious milk and a preparation method thereof. The DHA-containing watermelon juice nutritious milk provided by the invention comprises the following components according to the parts by weight: 30-58 parts of milk, 15-60 parts of watermelon juice and 0.4-3.0 parts of microalgae DNA powder. The preparation method of the DHA-containing watermelon juice nutritious milk comprises the following steps: mixing milk and microalgae DNA solution according to the ratio, adding watermelon juice, homogenizing, sterilizing and packaging. The DHA-containing watermelon juice nutritious milk has scientific formula, can strengthen the memory and thinking and improve intelligence, and has high nutritional value and simple preparation technology.
Owner:BEIJING JIANJIAN KANGKANG BIO TECH +1
Method for preparing barium tungstate nanometer double-line arrays by using DNA as templates
InactiveCN101805022AMorphological rulesUniform sizeNanostructure manufactureTungsten compoundsEscherichia coliDNA Solutions
The invention discloses a method for preparing barium tungstate nanometer double-line arrays by using colon bacillus genome DNA as templates, which belongs to the technical field of nanometer materials. The method comprises the following steps: adding a barium nitrate solution into a colon bacillus genome DNA solution; uniformly mixing the solution; carrying out oscillation hatching for 48 to 72 h under the conditions of the temperature between 4 and 6 DEG C and the oscillation speed between 80 and 90 r / min; and then, adding the sodium tungstate solution for oscillation hatching for 48 to 72 h under the conditions of the temperature between 4 and 6 DEG C and the oscillation speed between 80 and 90 r / min; . When the mixed solution is heated for 6 to 8 h under the condition of the temperature between 80 and 85 DEG C, the barium tungstate nanometer double-line arrays using the colon bacillus genome DNA as templates can be obtained. The invention avoids the complicated preparation process of the conventional method, can be completed under the mild conditions of low temperature, normal pressure and the like, the technology is simple, the cost is low, and the reaction can be easily controlled, so the goal of using the colon bacillus genome DNA as templates for synthesizing the barium tungstate nanometer double-line arrays to construct nanometer devices can be realized.
Owner:YANSHAN UNIV
Method for determining melamine content by using ultraviolet spectroscopy
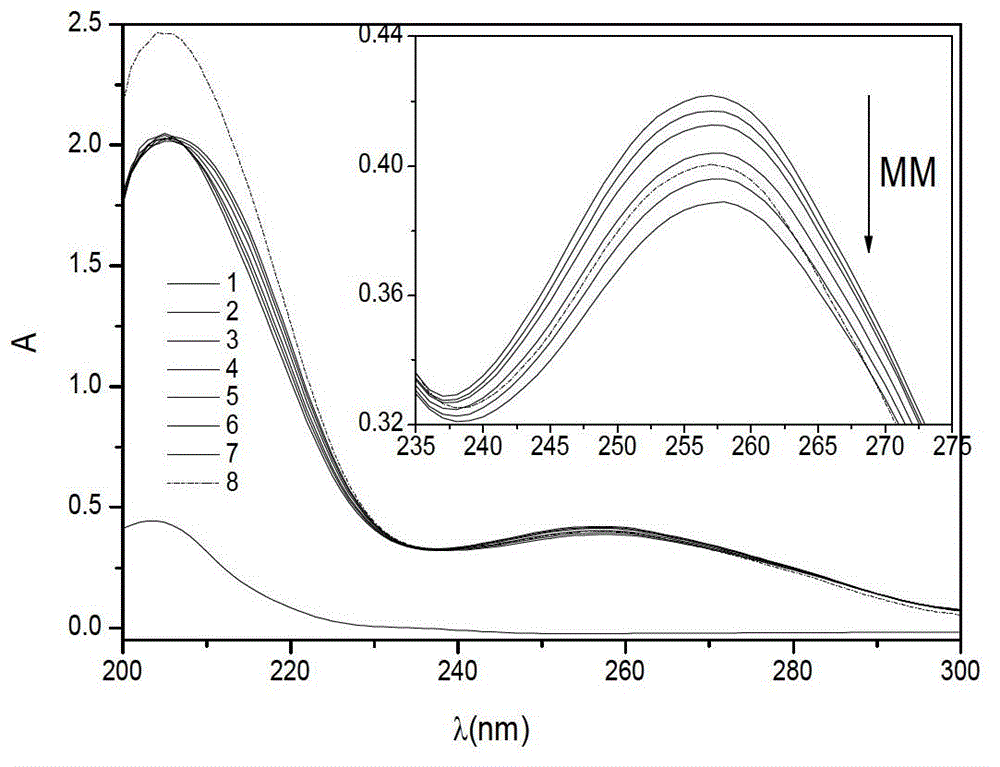
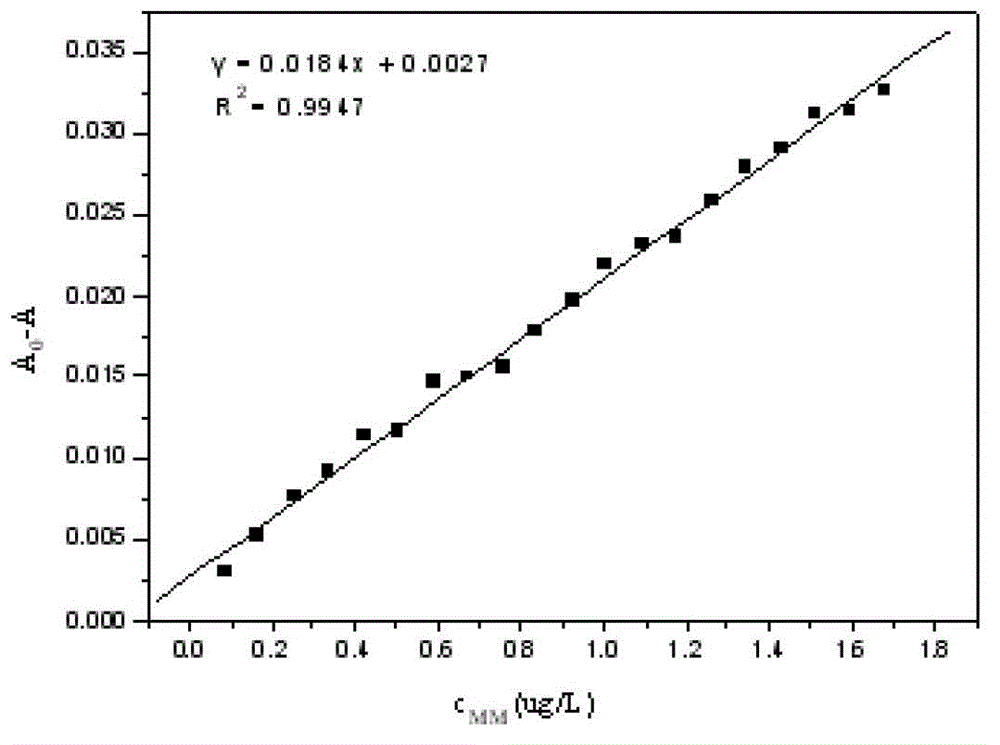
InactiveCN102749301AAccurate and effective determinationEasy to operateColor/spectral properties measurementsDNA SolutionsUv spectrum
The invention discloses a method for determining a melamine content by using ultraviolet spectroscopy. The method comprises steps of preparing a Tris-HCl buffer solution with a pH value of 4 to 10; preparing a melamine solution of 0 to 10 [mu]g / L, a DNA solution of 1.0 x 10<-5> to 1.5 x 10<-4> mol / L and a mixing solution of the melamine and DNA by using the Tris-HCl buffer solution, and determining absorbance of each solution using a blank reagent as a reference; establishing a working curve using a melamine concentration as an abscissa, and using differences between the DNA absorbance and the absorbance of the mixing solution of the melamine and the DNA as an ordinate; and adding a to-be-measured melamine liquid into the DNA solution, determining the absorbance, calculating the melamine concentration in accordance with the working curve, and converting the concentration into a corresponding melamine content in the to-be-measured liquid. The method employs an ultraviolet spectrum detection technology established on the basis of interactions of the melamine and deoxyribonucleic acid, and is accurate, simple, convenient and repeatable.
Owner:NINGBO INST OF TECH ZHEJIANG UNIV ZHEJIANG
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com