Bacterial cell computing component that indicates priority of a signal at different accompanying signals
A technology for indicating signals and computing components, which is applied in the field of synthetic biology and can solve problems such as incomparability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
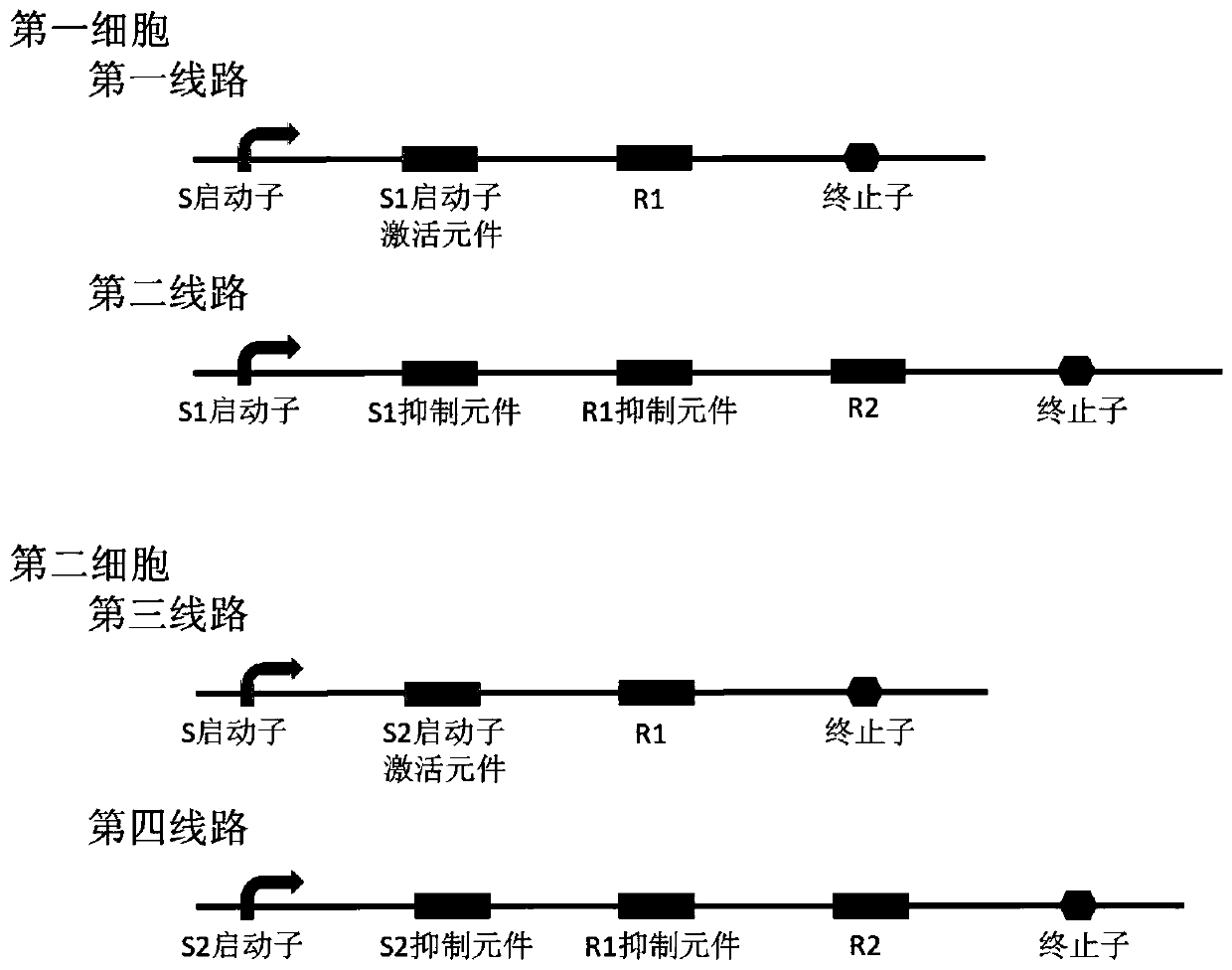
[0044] The signal S to be detected, the first accompanying signal S1 , the second accompanying signal S2 , the first indication signal R1 , and the second indication signal R2 are shown in the table below.
[0045] Signal to be detected S Ara (arabinose) First accompanying signal S1 IPTG (isopropyl-β-D-thiogalactopyranoside) Second accompanying signal S2 aTc (Anhydrotetracycline) The first indication signal R1 Green fluorescent protein GFP (that is, green light means 0) The second indication signal R2 Red fluorescent protein RFP (that is, red light indicates 1)
[0046] The promoter is specifically: the S promoter adopts the promoter P initiated by Ara BAD , the S1 promoter adopts the promoter P initiated by IPTG lac , the S2 promoter adopts the promoter P initiated by aTc tet .
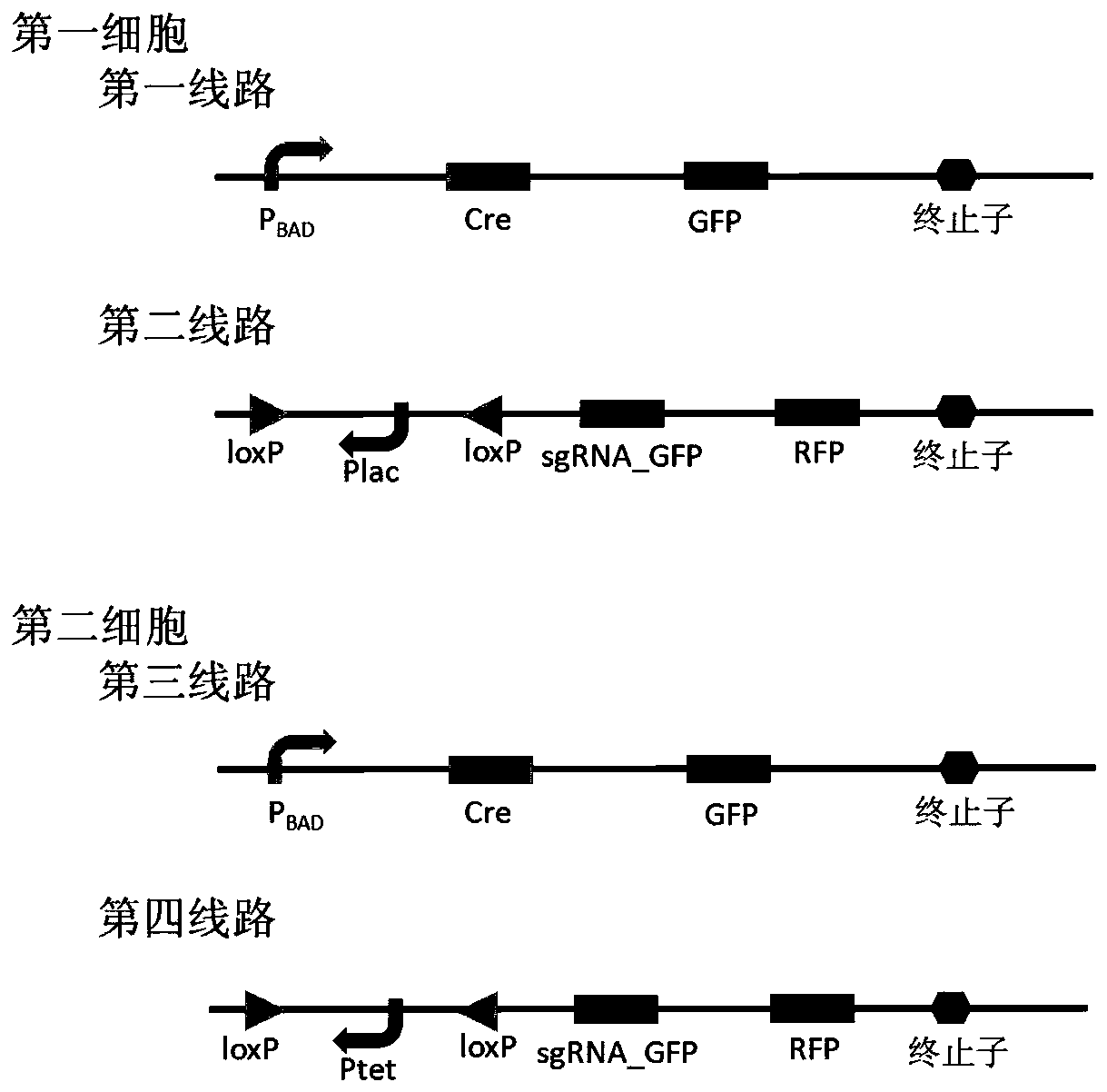
[0047] In this example, the CRISPR / dCas9 system is used to realize the indicator signal inhibition function. The dCas9 protein is a protein that loses th...
Embodiment 2
[0056] In this example, the CRISPR / dCas9 system in Example 1 is replaced with CRISPR / ddCpf1. The ddCpf1 protein is similar to the dCas9 protein and cannot cut DNA, but it can bind to DNA to prevent transcription from occurring. sgRNA is used to guide the ddCpf1 protein and the corresponding DNA fragment Binding, thereby preventing the occurrence of transcription and making the gene element unable to be expressed. E. coli strains or other strains that can ensure the work of CRISPR / ddCpf1 should be selected or constructed, and the sgRNA design can be in accordance with the requirements of the CRISPR / ddCpf1 system, and the others are the same as in Example 1.
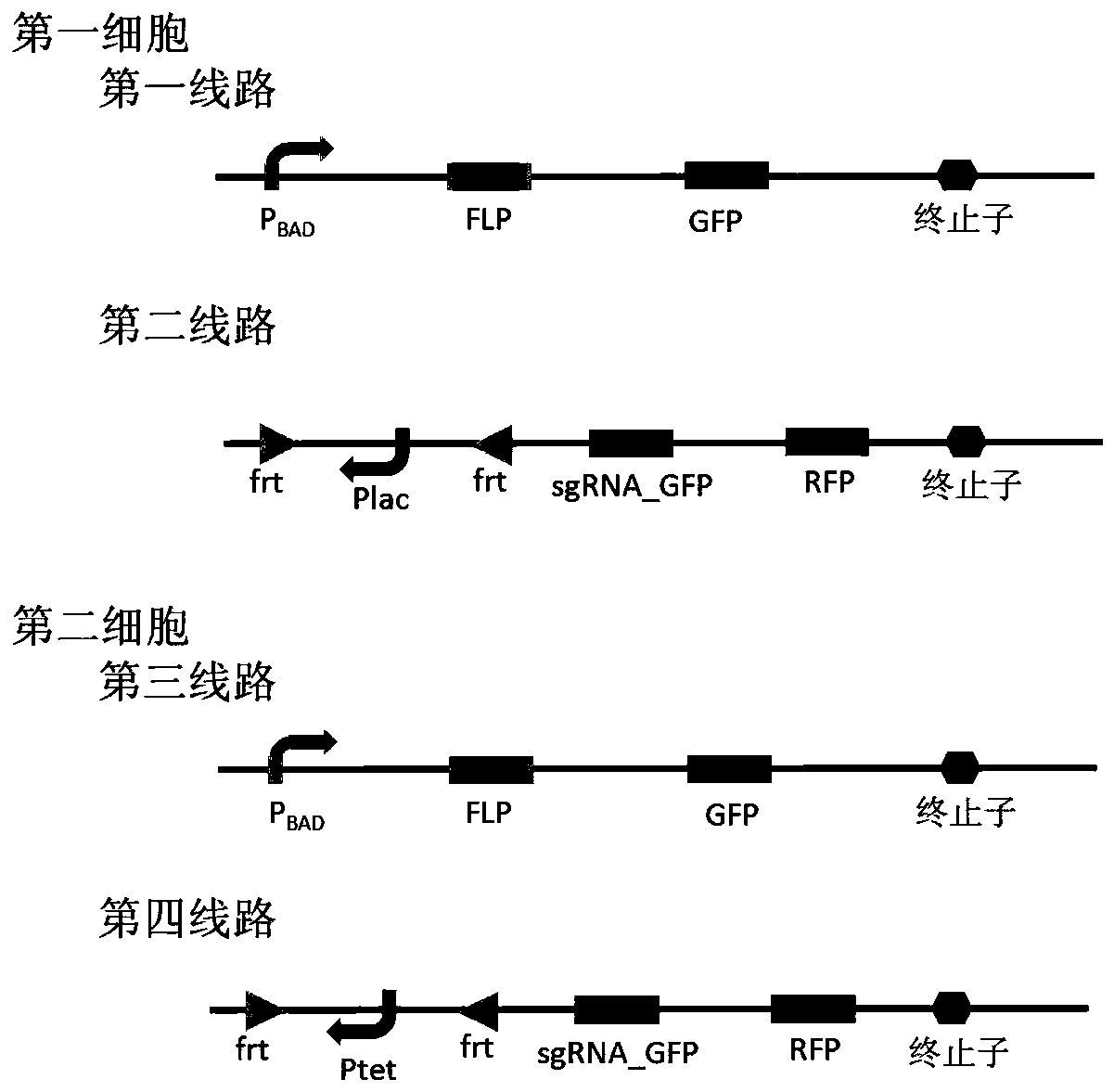
Embodiment 3
[0058] In this embodiment, the Cre recombinase and loxP sites in Example 1 are replaced with FLP recombinase and frt sites, such as image 3 , and others are the same as in Embodiment 1. The function of FLP recombinase is similar to that of Cre recombinase, and the frt site is its corresponding site. FLP recombinase can delete the fragment between two forward frt sites, and can also flip between two reverse frt sites. fragments.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com