A set of primers for identification of stored booklice and its application
A technology for storing booklice and booklice, which is applied in the direction of recombinant DNA technology, biochemical equipment and methods, and microbial measurement/inspection. Easy to operate and use, low cost, accurate and reliable identification results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Embodiment 1, the design of specific primer pair
[0073] The present invention designs specific primer pairs for eight common stored-grain booklices, as shown in Table 1, on the basis of obtaining the mitochondrial COI gene DNA barcode segments of ten common stored-grain booklices, and through multiple sequence alignment analysis.
[0074] Table 1 List of specific primers for 8 common storage booklice species in the world
[0075]
[0076]
Embodiment 2
[0077] Embodiment 2, the specific detection of primer
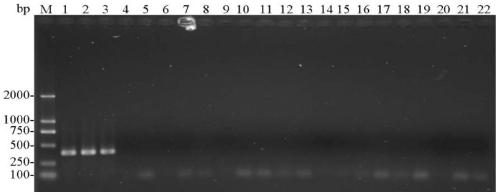
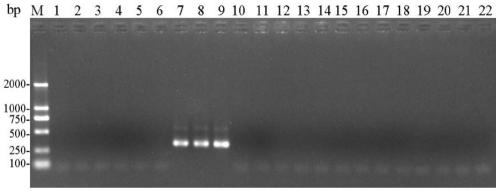
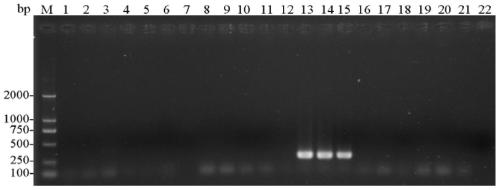
[0078] 1. Specific detection of entoF3 and entoR2
[0079] (1) Using the CTAB method to extract the genomic DNA of a single (or multiple) head of the stored booklice from sample 1 to sample 21.
[0080] (2) Use each genomic DNA obtained in step (1) as a template, and use entoF3 and entoR2 as primers to carry out PCR amplification to obtain each PCR amplification product, and simultaneously use ddH 2 O replaced the template as a negative control.
[0081] PCR amplification system (total volume 25 μL): 2×Taq PCR MasterMix (purchased from Sangon Bioengineering (Shanghai) Co., Ltd.) 12.5 μL, ddH 2 O 9.5 μL, 1 μL of template with a concentration of 100-300 ng / μL, and 1 μL of primers with a concentration of 10 μM.
[0082] PCR amplification program: pre-denaturation at 94°C for 3 min; denaturation at 94°C for 30 sec, annealing at 50°C for 30 sec, extension at 72°C for 30 sec, 30 cycles; extension at 72°C for 5 min.
[0083]...
Embodiment 3
[0115] Embodiment 3, the sensitivity detection of primer
[0116] 1. Sensitivity detection of entoF3 and entoR2
[0117] (1) Genomic DNA of sample 2 was extracted by CTAB method and diluted to 0.01ng / μL, 0.1ng / μL, 1ng / μL, 5ng / μL, 10ng / μL, 25ng / μL, 50ng / μL, 75ng / μL μL and 100ng / μL concentrations.
[0118] (2) Using the genomic DNA of each dilution obtained in step (1) as a template, and using entoF3 and entoR2 as primers to carry out PCR amplification to obtain each PCR amplification product, and ddH2O instead of the template as a negative control.
[0119] The PCR amplification system is as in Example 2.
[0120] The PCR amplification procedure was as in Example 2.
[0121] (3) Each PCR amplification product is carried out to agarose gel electrophoresis, the result is as follows Figure 9 shown.
[0122] Figure 9 Middle, M: DNA relative molecular weight standard (D2000); 9 different concentrations of DNA in μL and 100ng / μL were PCR amplification products as templates; ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| body length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com