Primer group for identifying storage book lice and application of primer group
A technology for storing book lice and book lice, which is applied in the directions of recombinant DNA technology, microbial determination/inspection, biochemical equipment and methods, etc. Easy to operate and use, low cost, accurate and reliable identification results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Example 1. Design of specific primer pairs
[0073] The present invention designs specific primer pairs for eight common storage book lice based on the mitochondrial COI gene DNA barcode segment of ten kinds of common storage book lice, after multiple sequence comparison analysis, as shown in Table 1.
[0074] Table 1 List of 8 common storage book lice specific primers in the world
[0075]
[0076]
Embodiment 2
[0077] Example 2. Specific detection of primers
[0078] 1. Specific detection of entoF3 and entoR2
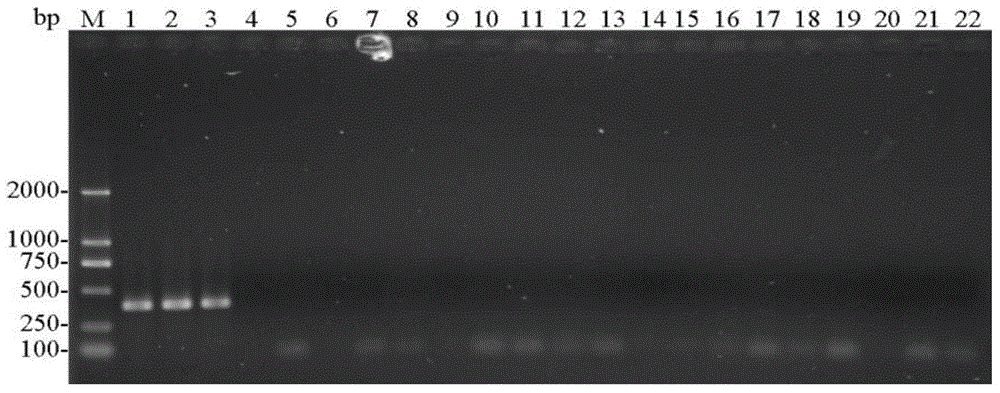
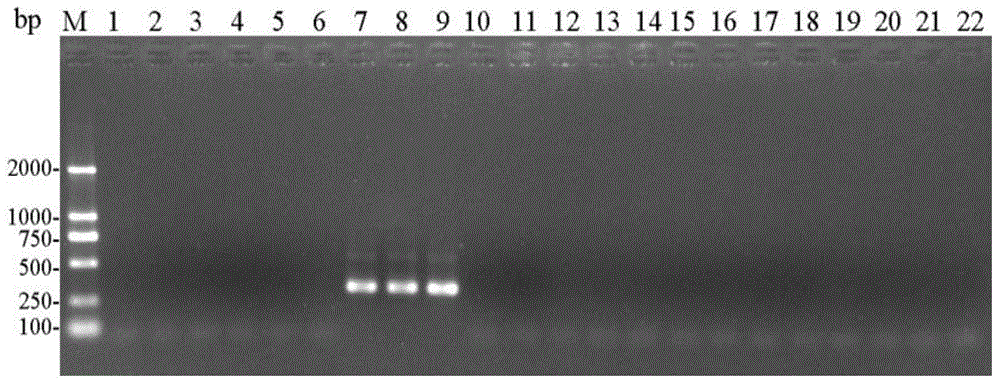
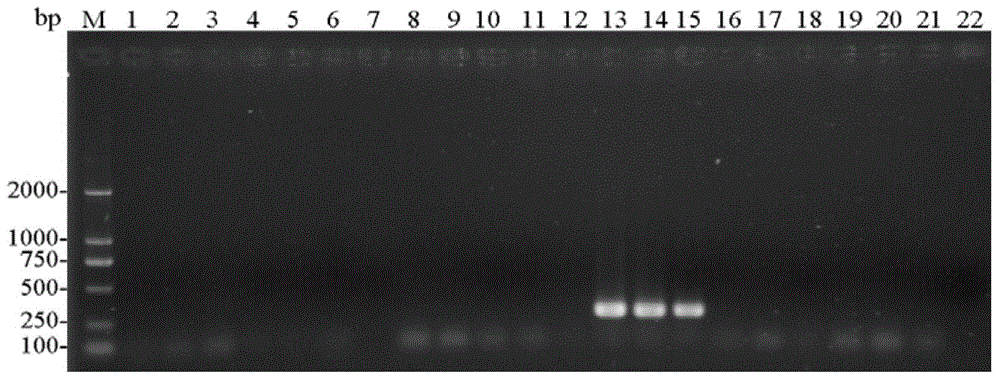
[0079] (1) Using the CTAB method to extract the genomic DNA of the single head (or multiple heads) of the storage book lice from sample 1 to sample 21 respectively.
[0080] (2) Use each genomic DNA obtained in step (1) as a template, and use entoF3 and entoR2 as primers for PCR amplification to obtain each PCR amplification product, and use ddH 2 O replaces the template as a negative control.
[0081] PCR amplification system (total volume 25μL): 2×TaqPCRMasterMix (purchased from Shenggong Bioengineering (Shanghai) Co., Ltd.) 12.5μL, ddH 2 O9.5μL, 1μL of template with a concentration of 100-300ng / μL, and 1μL of primers with a concentration of 10μM.
[0082] PCR amplification program: 94°C pre-denaturation for 3min; 94°C denaturation for 30sec, 50°C annealing for 30sec, 72°C extension for 30sec, 30 cycles; 72°C extension for 5min.
[0083] (3) Perform agarose gel electrophoresis on each ...
Embodiment 3
[0115] Example 3. Sensitivity detection of primers
[0116] 1. Sensitivity detection of entoF3 and entoR2
[0117] (1) Use CTAB method to extract the genomic DNA of sample 2 and dilute it to 0.01ng / μL, 0.1ng / μL, 1ng / μL, 5ng / μL, 10ng / μL, 25ng / μL, 50ng / μL, 75ng / μL and 100ng / μL concentration.
[0118] (2) Using the genomic DNA of each dilution obtained in step (1) as a template, and using entoF3 and entoR2 as primers for PCR amplification, each PCR amplification product is obtained, and ddH2O is used as a negative control instead of the template.
[0119] The PCR amplification system is as in Example 2.
[0120] The PCR amplification procedure is as in Example 2.
[0121] (3) Perform agarose gel electrophoresis on each PCR amplified product, and the result is as follows Picture 9 Shown.
[0122] Picture 9 Among them, M: DNA relative molecular weight standard (D2000); 1 to 9 are 0.01ng / μL, 0.1ng / μL, 1ng / μL, 5ng / μL, 10ng / μL, 25ng / μL, 50ng / μL, 75ng / μL and 100ng / μL 9 different concentratio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com