Reagent and application for detecting mutation of forkhead transcription factor o1 gene
A technology for detecting reagents and reagents, applied in the field of reagents for the detection of forkhead transcription factor O1 gene mutations, can solve the problems of reducing and affecting Wnt signal pathway conduction, and achieve the effects of simple experimental operation, improved detection specificity, and inhibited amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
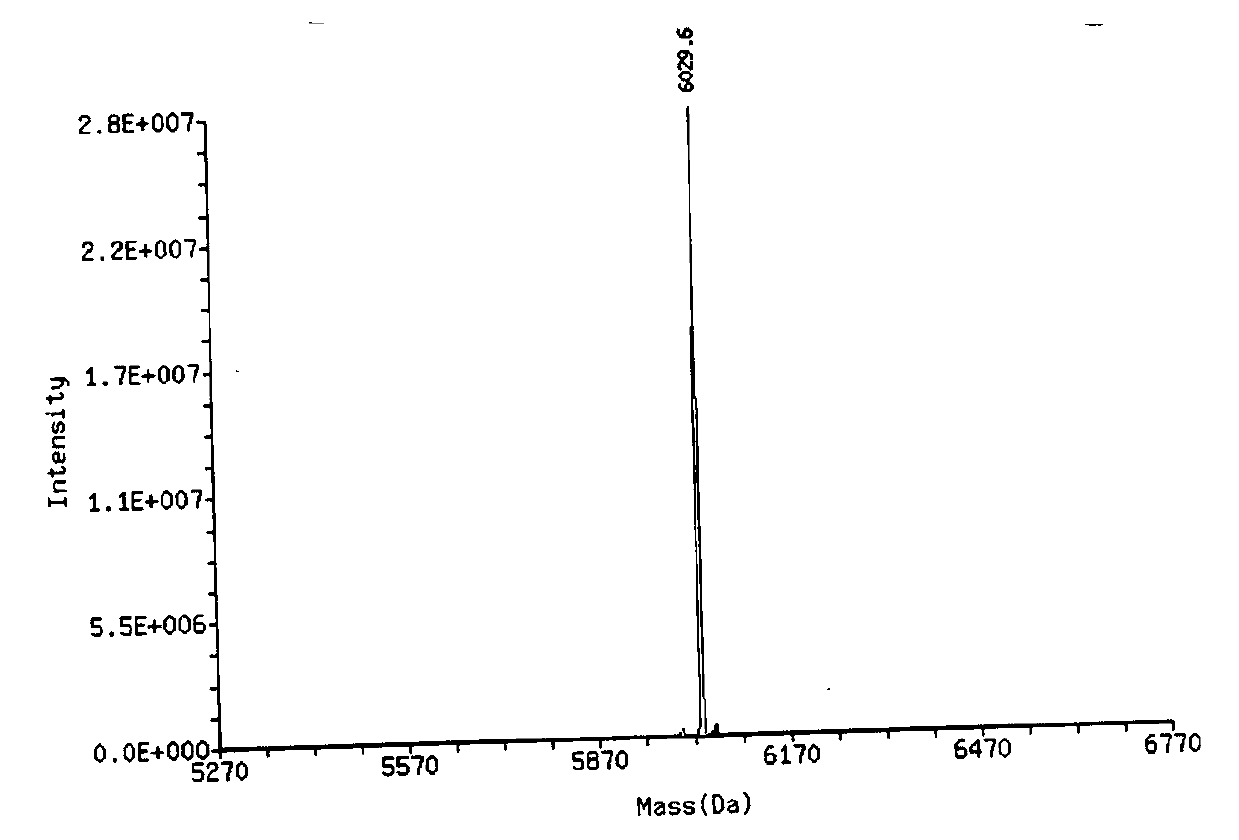
[0038] [Example 1] Primer probe design
[0039] According to the FOXO1 mRNA sequence (NCBIReference Sequence: NM_002015.3) reported by the National Center for Biotechnology Information (NCBI) (http: / / www.ncbi.nlm.nih.gov), using Applied Biosystems PrimerExpress Software for Real-Time PCR software design specific FOXO1 primers and probes.
[0040] FOXO1 R609C:
[0041] FOXO1-F:5'-CCTTTCTCCACCAGGAGAAGCTC-3'; (SEQ NO.1)
[0042] FOXO1-R:5'-CTTGACACTGTGTGGGAAGCTT-3'; (SEQ NO.2)
[0043] Mutant FOXO1(PNA)-P: 5'FAM-TGTTCATTGAGTGCTTAGAC-BHQ 3'; (SEQ NO.5)
[0044] Wild-type FOXO1-PNA: 5'-TGTTCATTGAGCGCTTAG-3'; (SEQ NO.7)
[0045] Mutant target sequence:
[0046] 9()
[0047] Wild-type target sequence:
[0048] (SEQ NO. 10)
[0049] FOXO1 D624N:
[0050] FOXO1-F:5'-CCTTTCTCCACCAGGAGAAGCTC-3'; (SEQ NO.3)
[0051] FOXO1-R:5'-CTTGACACTGTGTGGGAAGCTT-3'; (SEQ NO.4)
[0052] FOXO1(PNA)-P: 5'FAM-AATGACCTCATGAATGGAGATA-BHQ 3'; (SEQ NO.6)
[0053] FOXO1-PNA: AATGACCTCATGGATGGAGA; (...
Embodiment 2
[0062] [Example 2] Construction of recombinant plasmids containing FOXO1 gene mutant and wild-type DNA fragments
[0063] 1. Human endometrial carcinoma Ishikawa cell line culture and passage
[0064] 1) Cell culture
[0065] All cell lines were cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA) and 10% fetal bovine serum (Invitrogen, Carlsbad, CA) at 37°C and 5% CO2.
[0066] 2) Cell passage
[0067] First, use a sterilized pipette to suck out the culture medium in the cell culture dish, add PBS buffer to wash twice, slowly add an appropriate amount of trypsin to the cells, wait until the cells become round, adjust the angle and the cells can move, then add 3ml of 10 DMEM medium with % fetal bovine serum, after repeatedly blowing gently, observe the cell morphology and count under the microscope, pass appropriate amount of cells to other sterilized culture dishes according to the content of cells in the culture dish, add 5ml of DMEM medium Then put into 5% CO2 incubat...
Embodiment 3
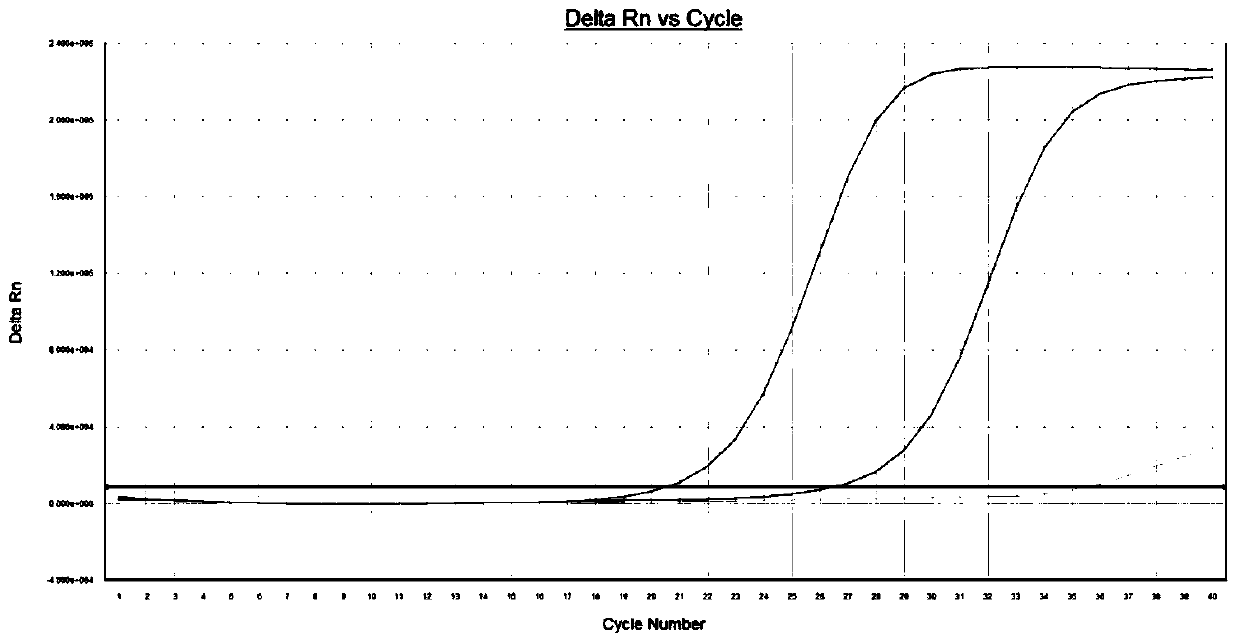
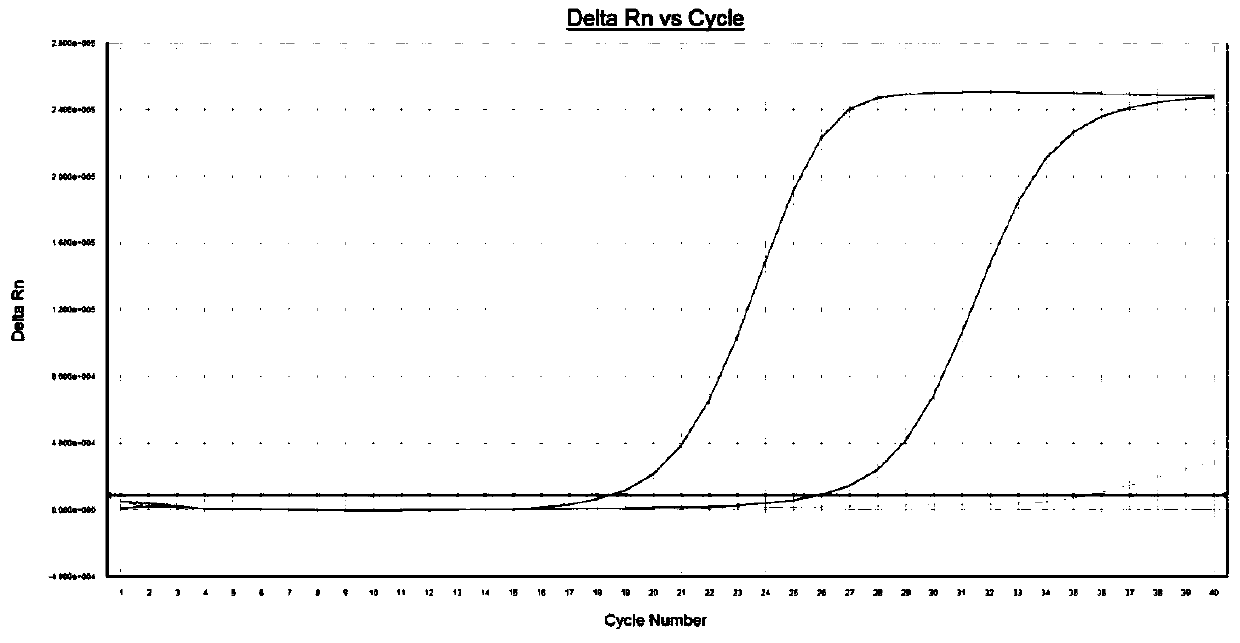
[0091] [Example 3] Amplified sample by real-time fluorescent quantitative PCR method
[0092] Take 1-5 μg of extracted total RNA and add it to the PCR reaction solution, which includes: sterile water, DNA polymerase with 5'→3' exolytic activity, dNTPs, 10×PCR Buffer, RNASIN, M-MLV reverse transcription Enzymes, oligo(dT) and solutions containing Mg ions. Among them, 0.3 μL of DNA polymerase with 5’→3’ exolytic activity at a concentration of 5 U / μL, 2 μL of dNTPs at a concentration of 10 mmol / L, 5 μL of 10×PCR Buffer, 0.6 μL of RNASIN at a concentration of 40 U / μL, and a concentration of 0.6 μL of M-MLV reverse transcriptase at 200 U / μL, MgCl at a concentration of 25 mmol / L 2 Solution 5 μL, add sterile water to a volume of 50 μL. Wherein, the DNA polymerase with 5'→3' excision activity can be Taq enzyme.
[0093] PCR amplification: put each reaction tube into the reaction tank of the fluorescent quantitative PCR instrument, set the type of labeled fluorescent group, sample n...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com