Methods for determining probe sets, kits and uses thereof
A probe set and kit technology, applied in the field of species abundance determination, can solve the problems of low probe efficiency, lack of exploration of heterologous sequence capture conditions, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
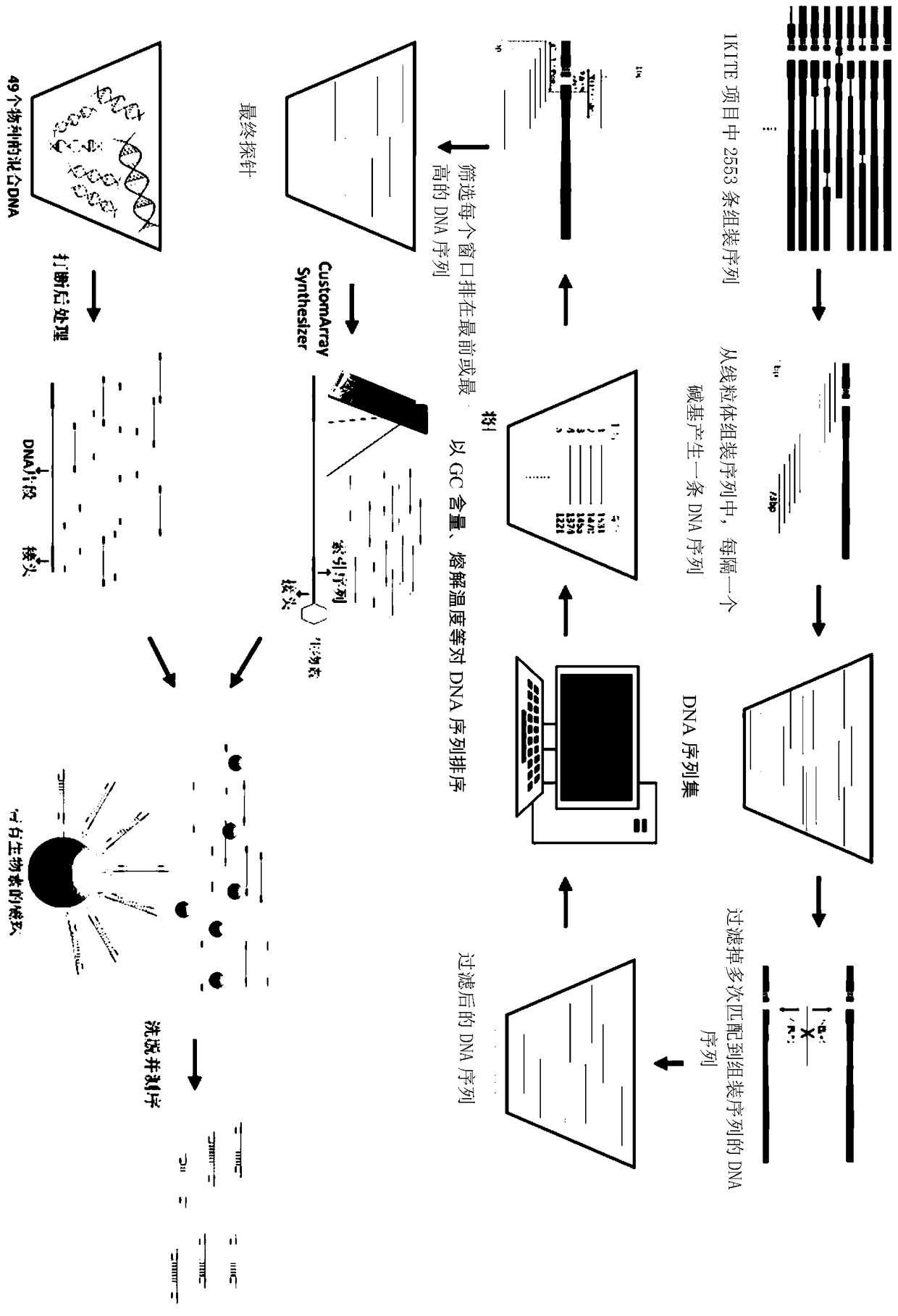
[0039] In order to ensure the coverage of the probes, the mitochondrial data from the 1KITE project were selected. The mitochondrial genome was obtained using the first 1KITE data of 379 species, and then the protein-coding genes on the mitochondria were obtained through gene annotation. Finally, 2,553 assembled sequences with an average length of 1,902bp were obtained for subsequent probe design. In the process of probe design, the mitochondrial assembly sequence was first divided into sequence sets with lengths of 64, 73 and 93, and the sequence set with a length of 73 bp was finally selected after comprehensive consideration of the theoretical melting temperature and relatively good mutation resistance, that is, the 1KITE The 2553 assembled sequences were segmented into 73bp long probes. All probes were then comprehensively ranked according to their specific region, theoretical melting temperature, and GC content. Divide all mitochondrial assembly sequences into 45bp wind...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com