Identification of predictive biomarkers associated with Wnt pathway inhibitors
A technology of biomarkers and inhibitors, applied in the field of cancer treatment, can solve problems such as uncontrolled proliferation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0288] Identify tumors that respond to the combination of OMP-18R5 and taxol (taxol)
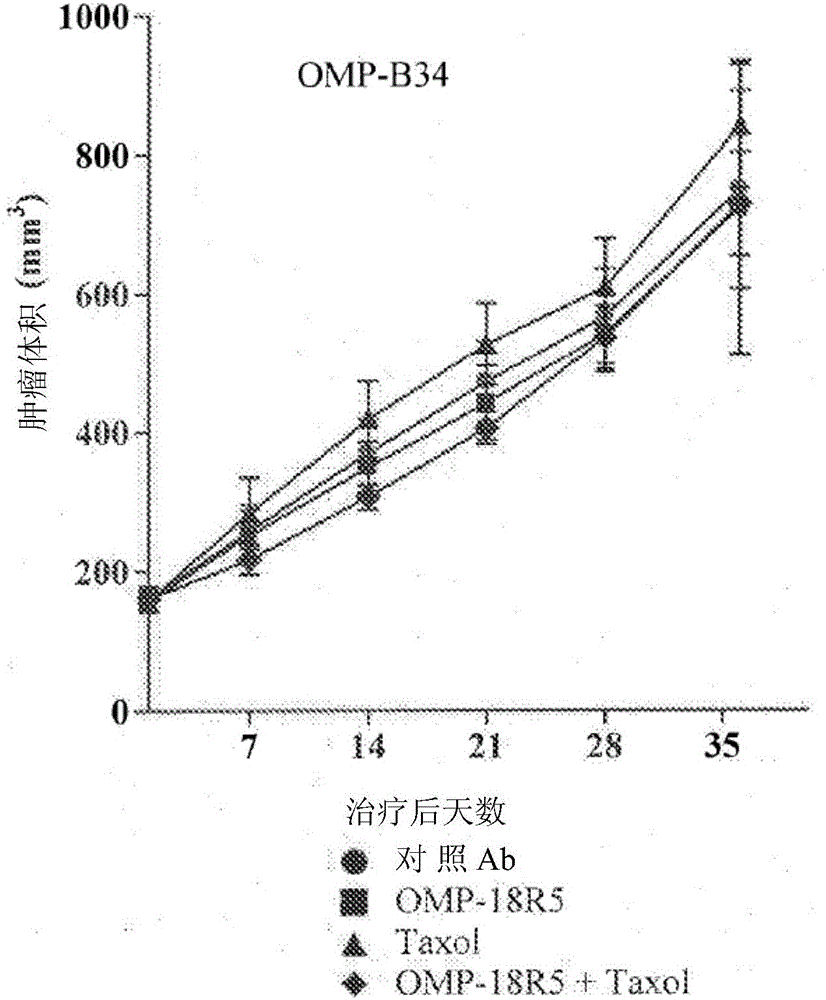
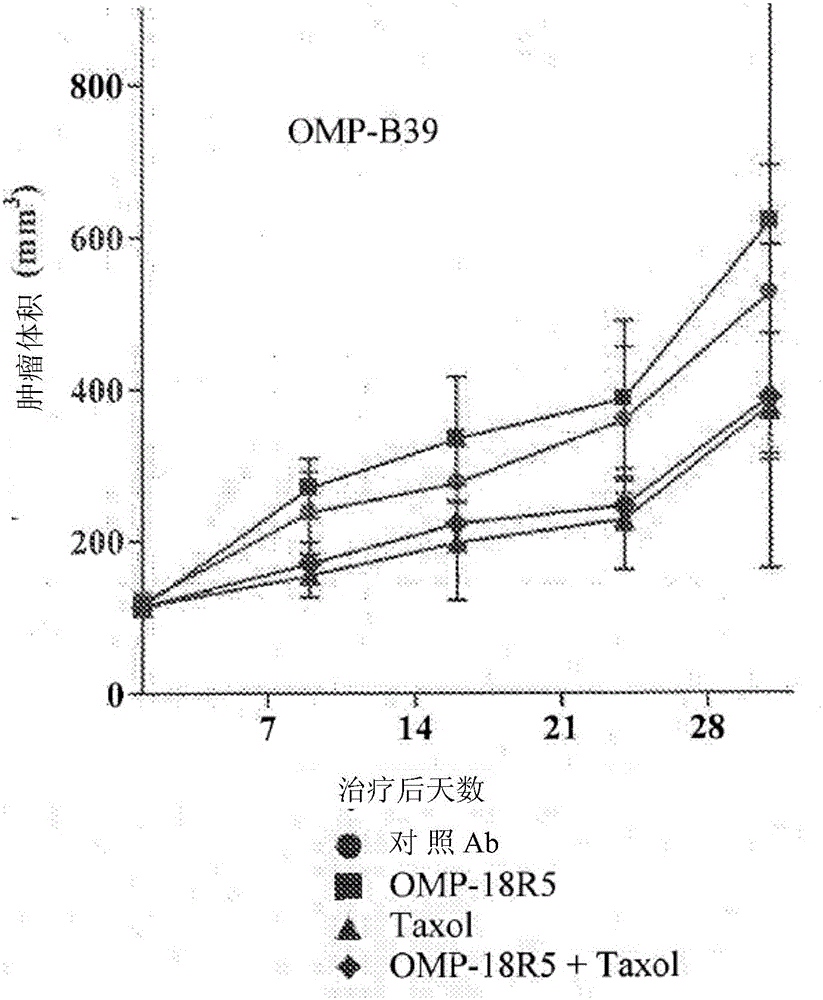
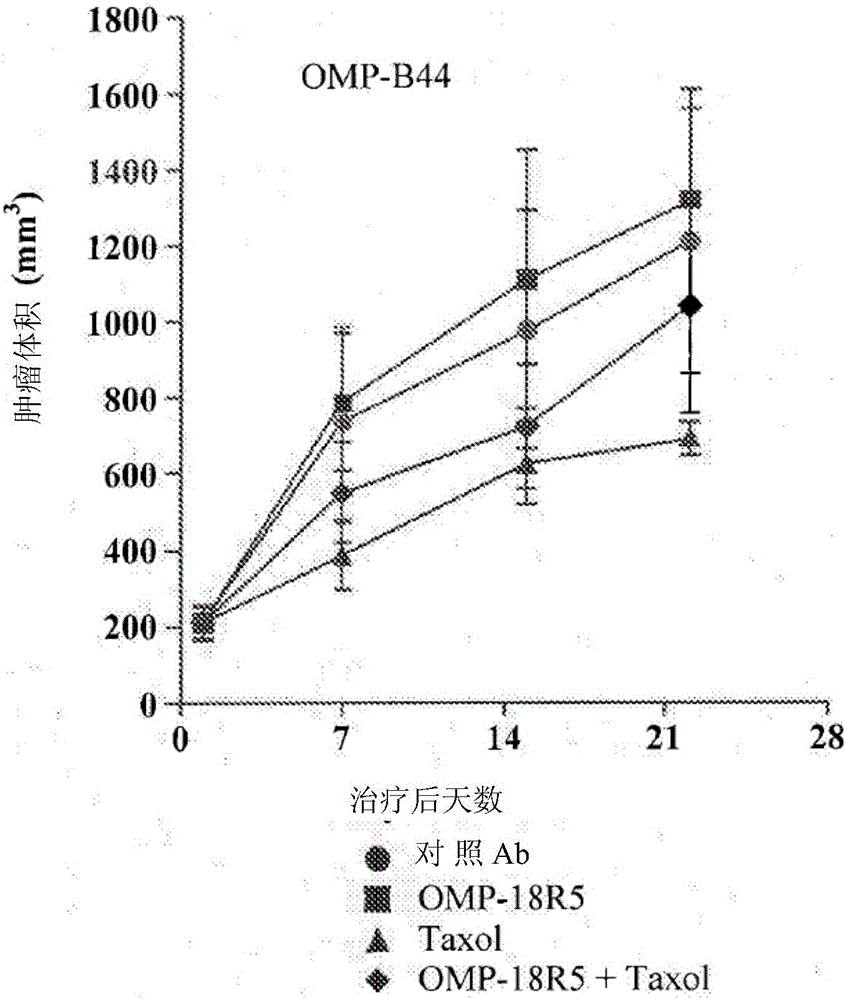
[0289] Breast tumor xenograft models OMP-B34, OMP-B39, OMP-B44, OMP-B59, OMP-B60, UM-T01, UM-T03 and UM-PE13 are derived from the smallest generation of tumor samples derived from patients. OncoMedPharmaceuticals or University of Michigan established. NOD / SCID mice aged six to eight weeks are injected subcutaneously with 2-4×10 4 One OMP-B34, OMP-B39, OMP-B44, OMP-B59, OMP-B60, UM-T01, UM-T03 or UM-PE13 tumor cell. Let the tumor grow until the tumor reaches an average volume of 100 to 150 mm 3 . Tumor mice were randomly divided into four groups (n=10 in each group) and were treated with control antibody 1B711 (15mg / kg), anti-FZD antibody OMP-18R5 (15mg / kg), paclitaxel (10mg / kg) or OMP-18R5 (15mg / kg) and paclitaxel (10mg / kg) combination therapy. Antibody and / or paclitaxel treatment is administered weekly. The tumor growth was monitored and the tumor volume was measured with electronic calipe...
Embodiment 2
[0295] Identify predictive biomarkers
[0296] The microarray analysis was based on untreated breast tumors OMP-B34, OMP-B39, OMP-B44, and UM-T01 that did not respond to the combination treatment of OMP-18R5 and paclitaxel ("non-responders"). The combination of 18R5 and paclitaxel was performed on untreated tumors OMP-B59, OMP-B60, UM-T03, and UM-PE13 that responded ("responders"). The RNeasy fibrous tissue mini kit (Qiagen, Valencia CA) and DNase treatment were used according to the manufacturer's instructions to isolate RNA from each tumor. The samples are stored at -80°C. The RNA was inspected on the Agilent 2100 bioanalyzer, and the integrity can be verified by the presence of intact 28S and 18S ribosomal peaks. All RNA samples have a 260 / 280 ratio> 1.8. The total RNA isolated from each tumor was amplified by Ovation RNA Amplification System V2 (NuGEN, San Carlos, CA). The amplified antisense single-stranded cDNA was segmented and biotinylated using FL-OvationcDNA biotin ...
Embodiment 3
[0306] In vivo validation of predictive biomarkers
[0307] Six additional breast cancer tumor lines were selected from the OncoMed tumor bank and were subjected to microarray analysis as described in Example 1. The six breast cancer tumors are OMP-B29, OMP-B71, OMP-B84, OMP-B90, UM-T02 and UM-T06. As described herein, the classification probability analysis using 6-gene biomarker labels is used to predict the response of each of these tumors to the combination of anti-FZD antibody OMP-18R5 and paclitaxel (see Figure 5 ). At the same time, the six tumors were evaluated in the in vivo xenograft model as described in Example 1 (see Figure 6A To F). As described in Example 1a, the "responders" in the in vivo model showed tumor growth inhibition compared with paclitaxel as a single agent, and the combination of OMP-18R5 and paclitaxel showed significantly higher tumor growth inhibition. The prediction system based on the classification probability is compared with the results of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com