Method for identifying a biomarker indicative of a reduced drug response using a thermal shift assay
A biomarker and drug technology, applied in biological testing, biomaterial analysis, measuring devices, etc., can solve problems such as technical proximity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0173] Materials and Methods
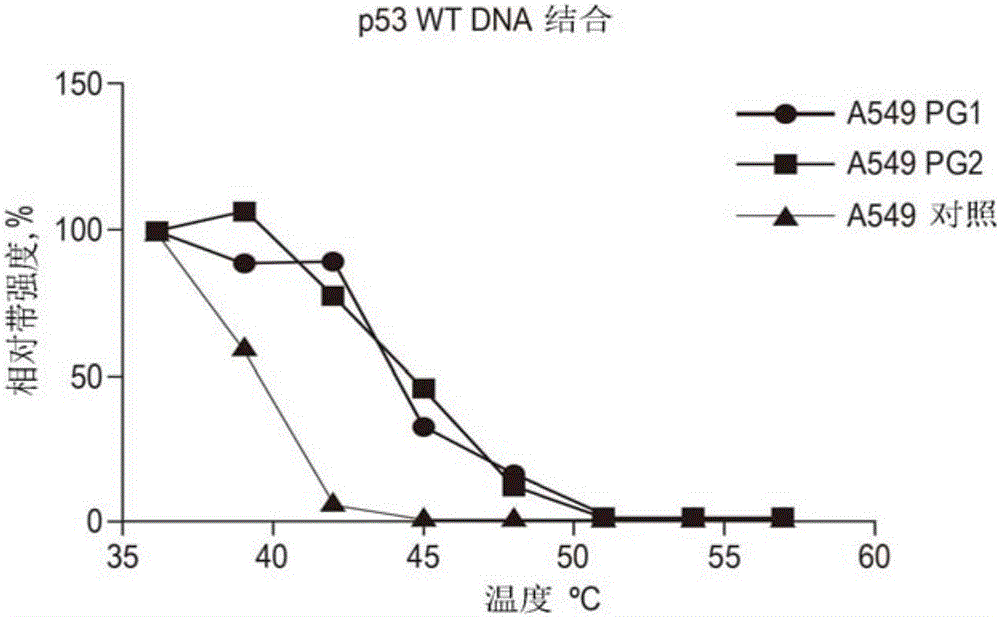
[0174] Human cancer cell line A549 (ATCC number CCL-185) was cultured in RPMI-1640 medium (Sigma-Aldrich) until it was cultured in a culture room (containing 5% CO 2 ), the RPMI-1640 medium contained 0.3g / L L-glutamine and was supplemented with 10% fetal bovine serum (FBS, Gibco / Life Technologies, Carlsbad, CA, USA ), 100 units / mL penicillin and 100 units / mL streptomycin (Gibco / Life Technologies).
[0175] Cell lysates were prepared by harvesting the cells and suspending the cell pellet in phosphate-buffered saline (PBS) to a cell concentration of 100,000 cells / ml at N 2 Three cycles of freeze-thaw in (l), and lysed cells were cleared from cell debris by centrifugation. The clarified lysate was divided into three aliquots and supplemented with DNA oligonucleotides PG1 or PG2 dissolved in ultrapure water, and as a control an equal amount of water was added to the third aliquot.
[0176] After incubation for 30 minutes at room temperature, the t...
example 2
[0178] Materials and Methods
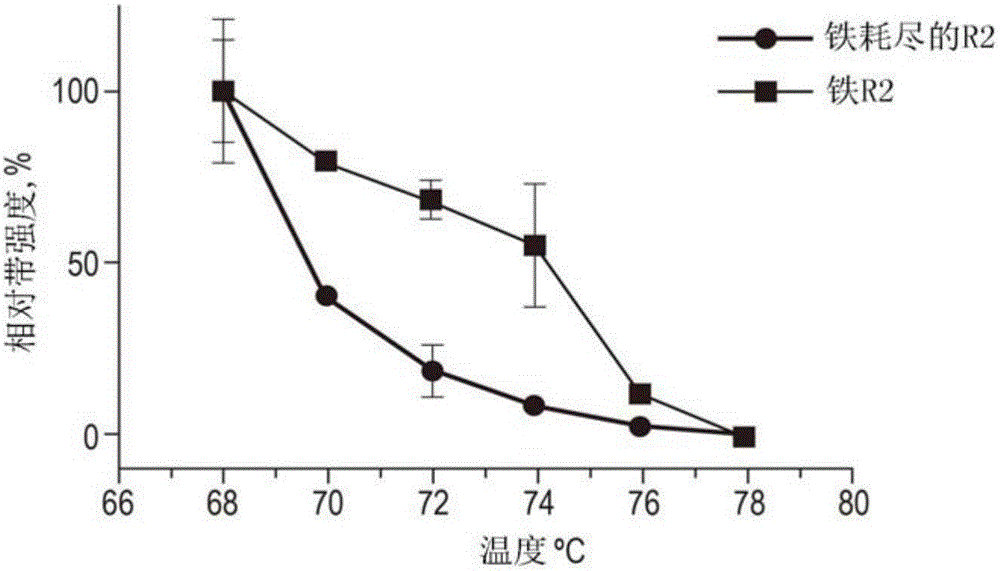
[0179] Cells were grown and lysed as described in Example 1. After clarification of the lysate, the supernatant was aliquoted and one was supplemented with 5 mM EDTA to a final concentration to chelate out metal ions. Control samples were supplemented with an equal volume of ultrapure water. After incubation for 10 minutes at room temperature, the lysates were aliquoted into 8 tubes of PCR strips and heated to increasing temperatures (68 to 78°C, using two degree increments) for a duration of three minutes. After heating, the samples were allowed to cool and the precipitated proteins were separated by centrifugation at 20000*g for 17 minutes. The remaining supernatants containing varying amounts of soluble protein were subjected to standard SDS-Page and Western blot analysis using a specific primary RNR R2 antibody (Santa Cruz Biotechnology SC-126) at a 1 :400 dilution in skim dietary milk. Using Graphpad Prism 6.0 software, the resulting prot...
example 3
[0181] Materials and Methods
[0182] Cells were grown and lysed as described in Example 1. After clarification of the lysate, the supernatant was divided into aliquots, one supplemented with a final concentration of 1 mM cyclic AMP dissolved in ultrapure water. Control samples were supplemented with an equal volume of ultrapure water. After incubation for 10 minutes at room temperature, the lysates were aliquoted into 8 tubes of PCR strips and heated to increasing temperatures (40 to 80° C., using four degree increments) for a duration of three minutes. After heating, the samples were allowed to cool and the precipitated proteins were separated by centrifugation at 20000*g for 17 minutes. The remaining supernatants containing varying amounts of soluble proteins were subjected to standard SDS-Page and Western blot analysis using specific first proteins against catalytic subunit α and regulatory subunit 1α at a 1:400 dilution in nonfat dietary milk Kinase A antibody (Santa C...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap