Molecular marker for identifying rice grain length characters, identification method and application
A molecular marker and molecular marker-assisted technology, applied in the field of molecular biology, can solve the problems of time-consuming, expensive operation, false positive, etc., and achieve the effect of low cost and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Using the above-mentioned molecular marker GS3-TPAP to identify the grain length traits of the tested rice materials, specifically includes the following steps:
[0032] The test materials were Minghui 63 and Nipponbare, which were the control materials in the GS3 gene cloning experiment. Minghui 63 carried the long-grain allele type GS3, and Nipponbare carried the short-grain allele type GS3.
[0033] S1, extract the genomic DNA of Minghui 63 and Nipponbare two rice materials to be tested respectively, and the genomic DNA extraction is carried out with the whole type gold plant DNA extraction kit (operate according to the kit instructions).
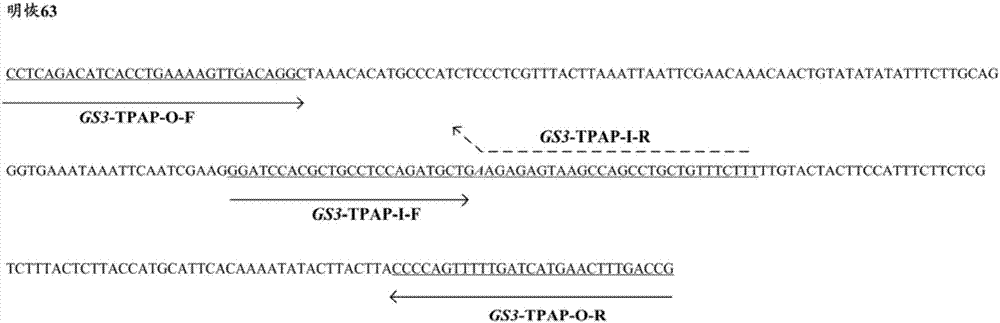
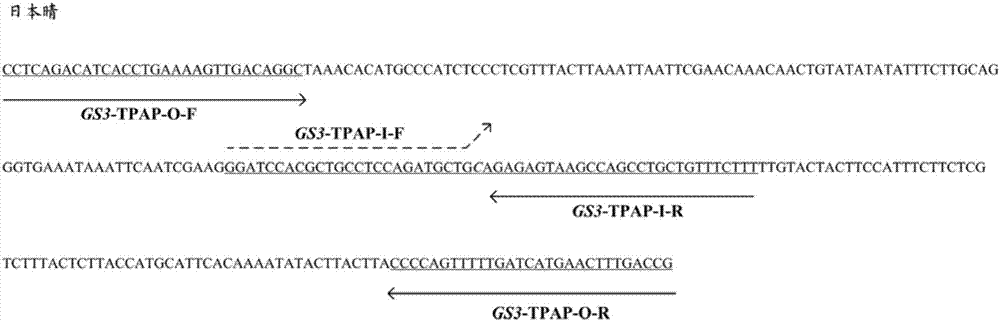
[0034] S2, use the genomic DNA obtained in S1 as a template, and perform PCR amplification with GS3-TPAP-O-F, GS3-TPAP-O-R, GS3-TPAP-I-F and GS3-TPAP-I-R as primers.
[0035] The PCR amplification reaction system is 10 μL: including 0.2 μL of DNA (1ng / μL); 0.3 μL each of GS3-TPAP-O-F and GS3-TPAP-O-R; Tap PCR Master Mix (with dye, ...
Embodiment 2
[0042] Genotype detection of offspring of DH population using molecular marker GS3-TPAP
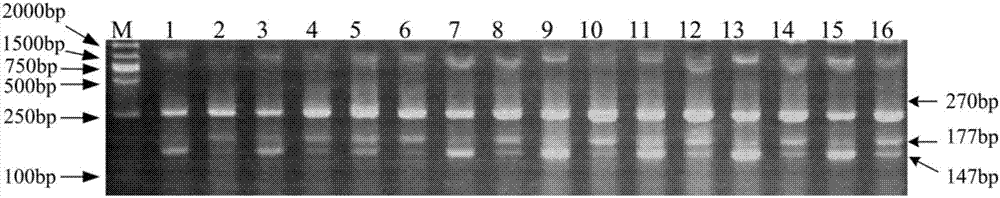
[0043] The DH population is a long-grain allele type rice variety Minghui 63 (male parent) hybridized with a short-grain allele type rice variety Qiuguang (female parent), 140 stable lines obtained from anther culture, and tested by marker GS3-TPAP The GS3 allelic genotypes of 140 stable lines were identified and compared with the long-grain phenotypes in the two years from 2012 to 2013. The results are shown in Table 2. In 2012, there were 59 long-grain allelic rice lines. The phenotypes are all long-grained, for example, the average grain length can reach 8.25 mm; there are 81 short-grain allelic rice varieties, and the phenotypes are all short-grained, such as the average grain length is only 5.56 mm; There are a total of 59 rice varieties with grain allelic type, all of which are long-grain phenotypes, for example, the average grain length can reach 8.44mm; there are 81 short-grain al...
example 2
[0047] Example 2 Genotype detection of offspring of RIL population using molecular marker GS3-TPAP
[0048] The RIL population is a long-grain allele type rice variety Minghui 63 (male parent) hybridized with a short-grain allele type rice variety Qiaoke 951 (female parent). 110 stable lines were obtained by single-grain transmission. GS3-TPAP identified the GS3 allele types of 110 stable lines, and compared them with the grain length phenotypes of the two years from 2013 to 2014. The results are shown in Table 3. In 2013, the long-grain allele type rice varieties There are 38 rice varieties in total, all of which are long-grained, with an average grain length of up to 8.38mm; a total of 72 short-grain allelic rice varieties, all of which have short-grained phenotypes, such as an average grain length of only 5.32mm; In 2014, there were a total of 38 long-grain allelic rice varieties, all of which were long-grain, with an average grain length of 8.39 mm; a total of 72 short-gra...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Length | aaaaa | aaaaa |
| Length | aaaaa | aaaaa |
| Length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com