Cross and progressive information extraction-based biological clustering method
A progressive extraction and clustering method technology, applied in informatics, biostatistics, bioinformatics, etc., can solve problems such as poor accuracy, loss of useful information, and inability to determine the type of biological clustering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
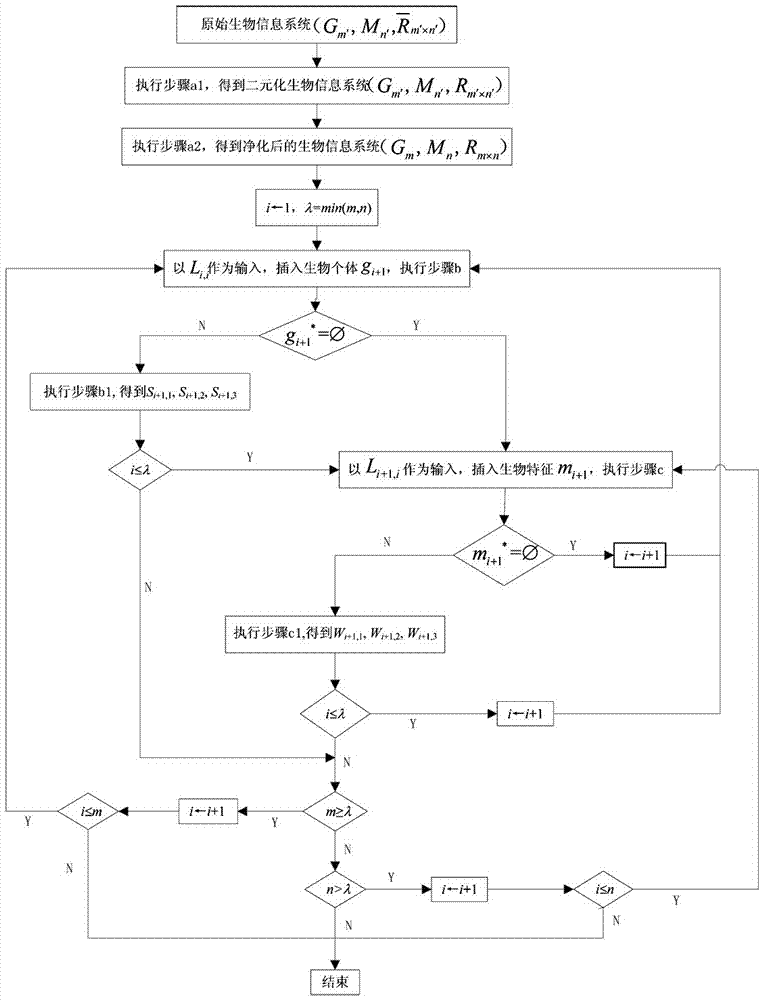
[0117] Given 10 weavers and their 9 biological characteristics constitute the original biological information system (see Table 1), that is, G 10 ={1,2,3,4,5,6,7,8,9,10}, M 9 ={a,b,c,d,e,f,g,h,i}, where a represents body length, b represents wing length, c represents wing width, d represents pronotum length, e represents pronotum Plate height, f represents the femur length of the forefoot, g represents the femur length of the midfoot, h represents the femur length of the hind foot, and i represents the length of the egg-laying valve; Set G for biological individuals 10 and biometric set M 9 The binary relationship between them is shown in Table 1.
[0118] Table 1 Original biological information system
[0119]
[0120] According to step a, the original biological information system Perform preprocessing to obtain the purified biological information system (G 5 , M 6 , R 5×6 ):
[0121] According to step a1, the original bioinformatics system To perform bin...
Embodiment 2
[0206] In order to further illustrate the practicability of the present invention, the data source used in this embodiment is the Katydid Research Laboratory of Hebei University. The laboratory personnel collected 300 spinosa insects from Guangxi, Guizhou, Tibet, Yunnan, Hainan, Jiangxi, Hunan, Zhejiang, Taiwan, Chongqing and other regions as experimental materials. The present invention extracts 57 female weavers as biological individual collection G 57 , and the biological characteristics of 6 kinds of textile mothers as the biological characteristic set M 6 . The six biological characteristics are: pronotum length to body length ratio (a), forefoot femur length to body length ratio (b), midfoot femur length to body length ratio (c), hind femur length Ratio to body length (d), ratio of oviposition petal length to body length (e), ratio of wing length to wing width (f). A collection of biological individuals G 57 with biometric set M 6 original biological information sys...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com