DNA methylation marker in DNA of host T cells and peripheral blood mononuclear cells (PBMCs) of cancer patients
A methylation marker and methylation technology, applied in the determination/testing of microorganisms, biochemical equipment and methods, etc., can solve unexplained problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
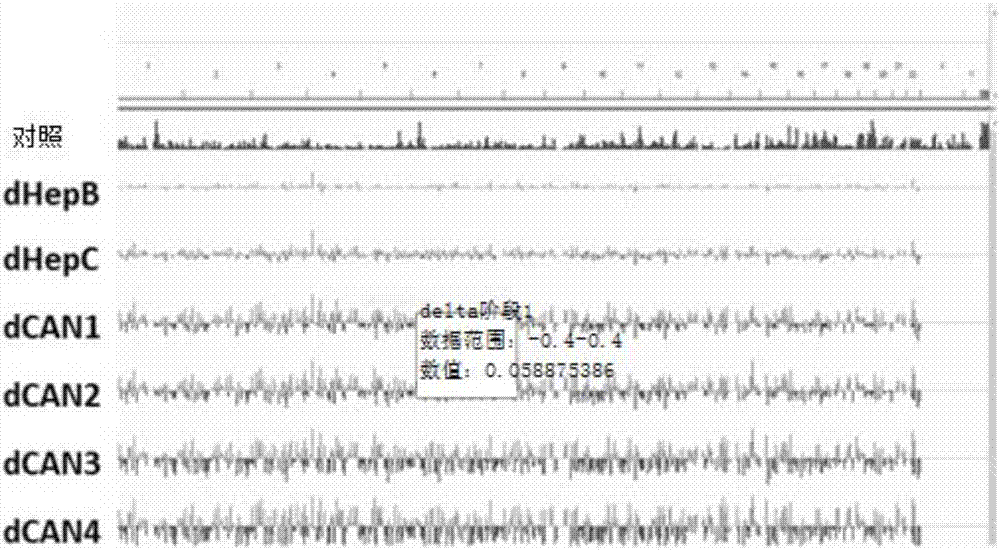
[0083] Example 1. DNA Methylation Markers in Peripheral Blood Mononuclear Cells (PBMCs) Associated with HCC Cancer Clinical Progression Stage
[0084] patient sample
[0085] HCC clinical progression stage was diagnosed according to the European Association for the Study of the Liver and European Organization for Cancer Therapy and Research EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. Patients were divided into four groups, including BLC-0 (Stage 1), BLC-A (Stage 2), BLC-3 (Stage 3), and BLC-C / D (Stage 4). For the sake of simplicity, the above-mentioned phases refer to phases 1 to 4 in the drawings and embodiments of the present invention. Confirm the diagnosis of chronic hepatitis B according to the American Association for the Study of Liver Diseases AASLD practice guidelines for hepatitis B, and the AASLD recommendations for the diagnosis of chronic hepatitis C, and testing, management, and treatment of hepatitis C. Strict exclusion cri...
Embodiment 2
[0100] Example 2. Unique and overlapping differential methylation sites are associated with different HCC clinical progression stages, and can distinguish HCC from hepatitis B and hepatitis C
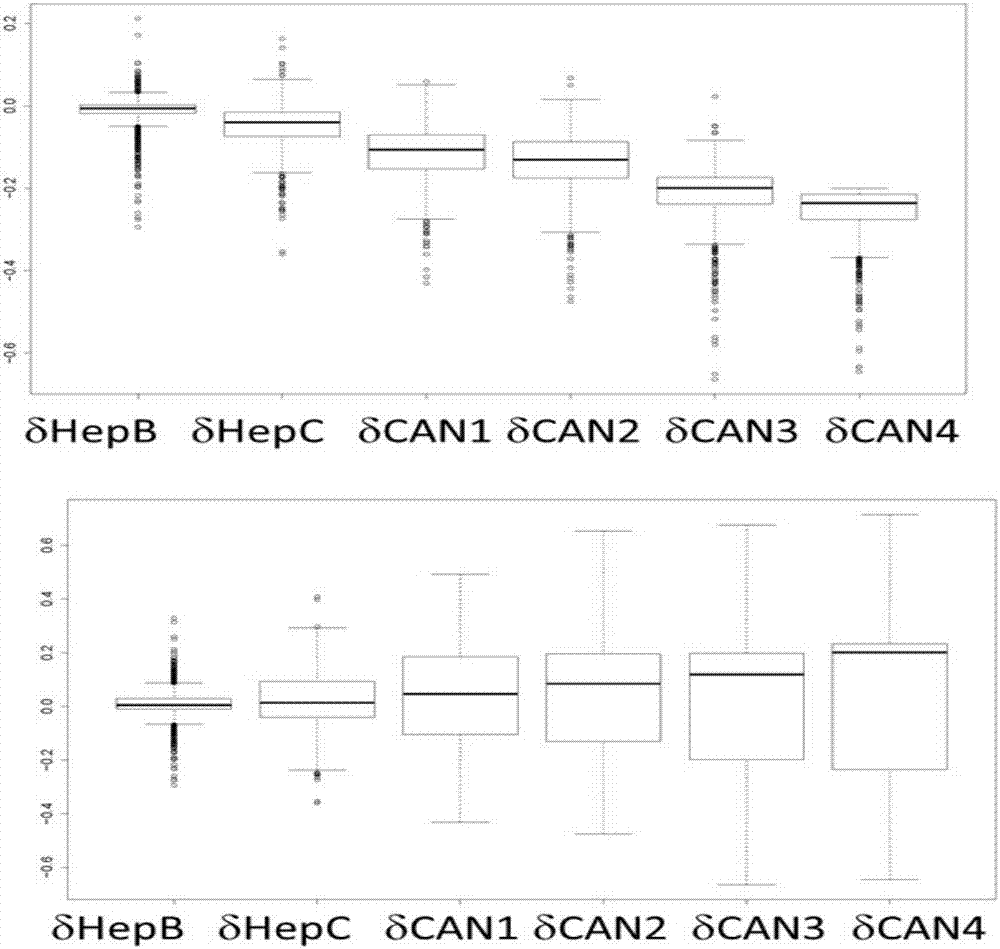
[0101] The inventors described differentially methylated CGs between healthy controls and each HCC clinical progression stage by using the implementation of the Bioconductor package Limma (50) in ChAMP. The number of differentially methylated CG sites between each HCC clinical progression stage and healthy controls (p-7 ) increased with the progress of clinical stage; stage 1 (BLC-0) was 14,375; stage 2 (BLC-A) was 22,018; stage 3 (BLC-B) was 30,709; stage 4 (BLC -Phase C / D) is 54580. The significance of the overlap between two groups was determined using R-based hypergeometric Fisher's exact test. Significant overlap exists between cancer stages ( Figure 3A ), which implies that there are affected common markers in all stages of clinical progression of HCC (p-297 ).
[0102] The ra...
Embodiment 3
[0108] Example 3: Cancer clinical progression stage-specific DNA methylation markers are used to predict unknown samples from patients by minus-one Pearson cluster analysis, thereby detecting HCC cancers in early clinical progression stages and distinguishing early clinical progression stages of HCC cancer and chronic hepatitis
[0109] Differential methylation sites for each HCC clinical progression stage were obtained by comparing 10 healthy controls and 10 clinical progression stage-specific HCC patients. Other clinical progression stages and hepatitis B and C samples were not "trained" by these differentially methylated CGs ("training" was applied to the model to obtain differentially methylated sites), they served as "cross-validation" for unknown samples " set to address the following questions: First, can markers derived from a stage of clinical progression of the cancer correctly classify HCC samples that were not "trained" by these markers? Second, could the DNA meth...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com