Method for Structural Annotation and Alignment Assessment of Full-Length Transcripts
A technology for comparing results and transcripts, applied in genomics, instrumentation, proteomics, etc. It can solve the problems of redundant and unclear matchAnnot results, and achieve the effect of optimizing the display method.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
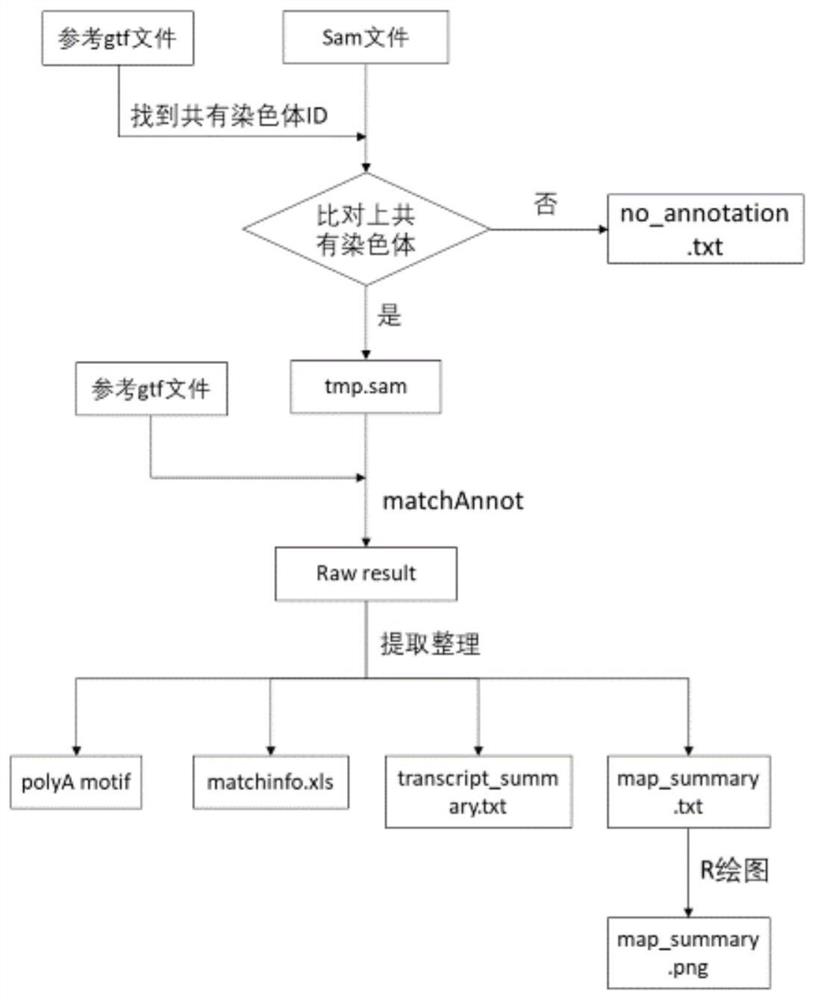
[0012] see figure 1 , the comparison result evaluation and gene structure annotation method, including the following steps:
[0013] (1) Obtain the chromosome ID shared by the reference genome annotation gtf file and the full-length transcriptome and reference genome comparison result sam file;
[0014] (2) Screen the entries of non-common chromosomes compared in the sam file, sort them out and output them to no_annotation.txt (this part of the full-length transcript can be supplemented with annotations without reference genome annotation to obtain new genes), and compared The entries of the shared chromosomes are output to tmp.sam;
[0015] (3) Use matchAnnot software for structural annotation and comparison result evaluation, tmp.sam file and gtf file as input files;
[0016] (4) Organize the matchAnnot results, output the polyA motif of the full-length transcript separately to polyA_motif.txt, extract the best matching reference gene and reference transcript information f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com