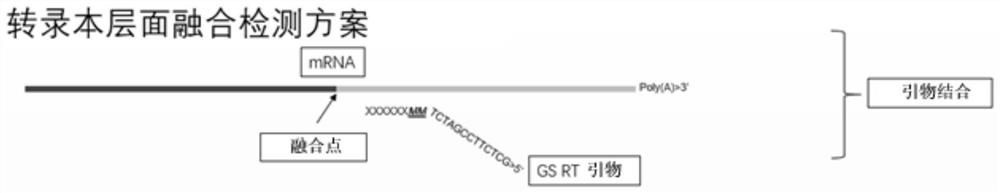
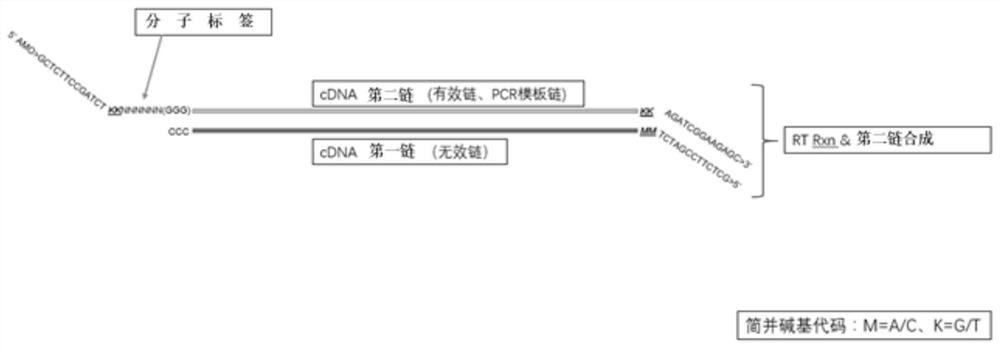
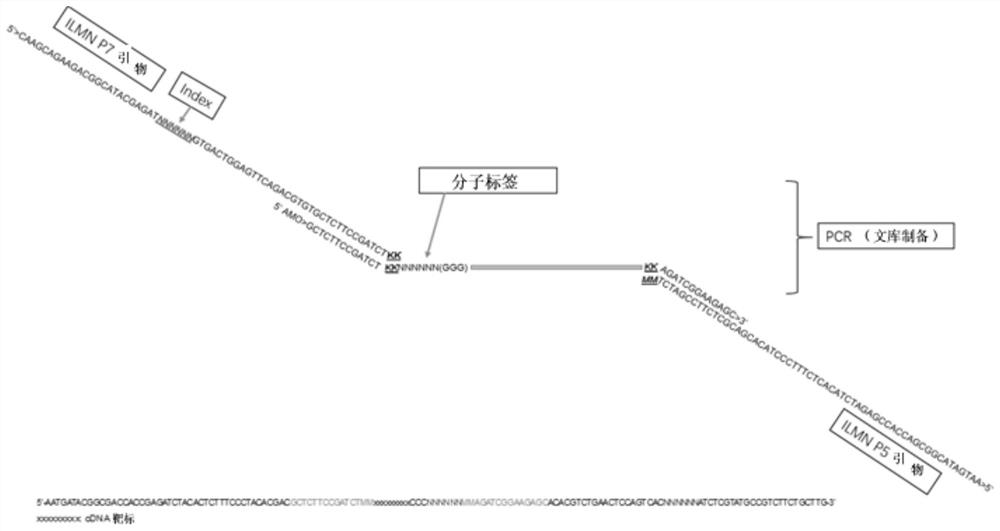
A method for detecting eml4-alk, ros1 and ret fusion gene mutations
A gene and unfused technology, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of poor detection specificity, large difference in result interpretation, low sensitivity, etc., to achieve high specificity and avoid pollution effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0087] Example 1 Detection sample processing and RNA extraction
[0088] 1. Test samples include fresh pathological tissue, frozen pathological sections, paraffin-embedded tissue or sections.
[0089] a. In the case of using cells as input, for fresh pathological tissue, first perform cell dispersion treatment, that is, grind the tissue in liquid nitrogen, add about 600 μL of PBS per 20-30 mg of tissue, and centrifuge at 12,000 rpm (~13,400 × g) After 1 minute, the cell pellet was obtained, the supernatant was discarded, the cell pellet was aspirated, and the cell suspension was diluted with 1x PBS to a density of ~500 cells / μL
[0090] b. In the case of using total RNA as input, take about 1 gram from fresh pathological tissue, frozen pathological sections, paraffin-embedded tissue or sections, and use Qiagen's RNA extraction kit to extract the RNA. Tissues were paraffin-embedded and their RNA was extracted using Qiagen's FFPE tissue RNA extraction kit. The above-mentioned ...
Embodiment 2
[0096] Example 2 Reverse transcription to obtain double-stranded full-length cDNA
[0097] 1. Pipette 1.7*N μL of RT Enzyme Mix into the RT Buffer of the previous step, mix by hand, and centrifuge;
[0098] 2. Add 15μL of RT Buffer and RT Enzyme Mix to each cell lysate, centrifuge briefly and place on ice immediately, and incubate the samples on a preheated PCR machine under the following conditions:
[0099]
[0100] Example 3 Amplification
[0101] Add 29.25 μL PCR Mix to the reverse transcription product (the solution volume is 49.25 μL at this time), add 0.75 μL Index primer to each reaction tube, mix well and centrifuge, and amplify in the PCR machine. The reaction conditions are as follows:
[0102]
Embodiment 4
[0103] Example 4 Library Detection
[0104] Step 1: Purification
[0105] 1) Transfer the amplification product to a centrifuge tube, take 0.8× (40uL) Ampure XP magnetic beads or CMpure magnetic beads, mix with the amplification product and place it on a magnetic stand for 10 minutes;
[0106] 2) After all the magnetic beads are adsorbed on the tube wall (about 5 min), discard the supernatant and wash the magnetic beads twice with newly prepared 80% ethanol, and discard the supernatant;
[0107] 3) Stand at room temperature for 5 minutes, and after the magnetic beads are dry (please be careful not to over-dry the magnetic beads to cause the magnetic beads to crack, so as not to affect the recovery efficiency), resuspend the magnetic beads with 17.5uL TE buffer, EB buffer or nucleic acid-free water according to downstream needs;
[0108] 4) After standing at room temperature for 5 minutes, place the centrifuge tube on a magnetic stand, and aspirate 15uL of the supernatant, whi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com