Multiplex nucleic acid assay methods capable of detecting closely related alleles, and reagents therefor
An assay method and multiplexing technology, which are applied in the fields of nucleic acid amplification and detection and determination, and can solve the problems of multiple assays without proof of multiplex assays.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
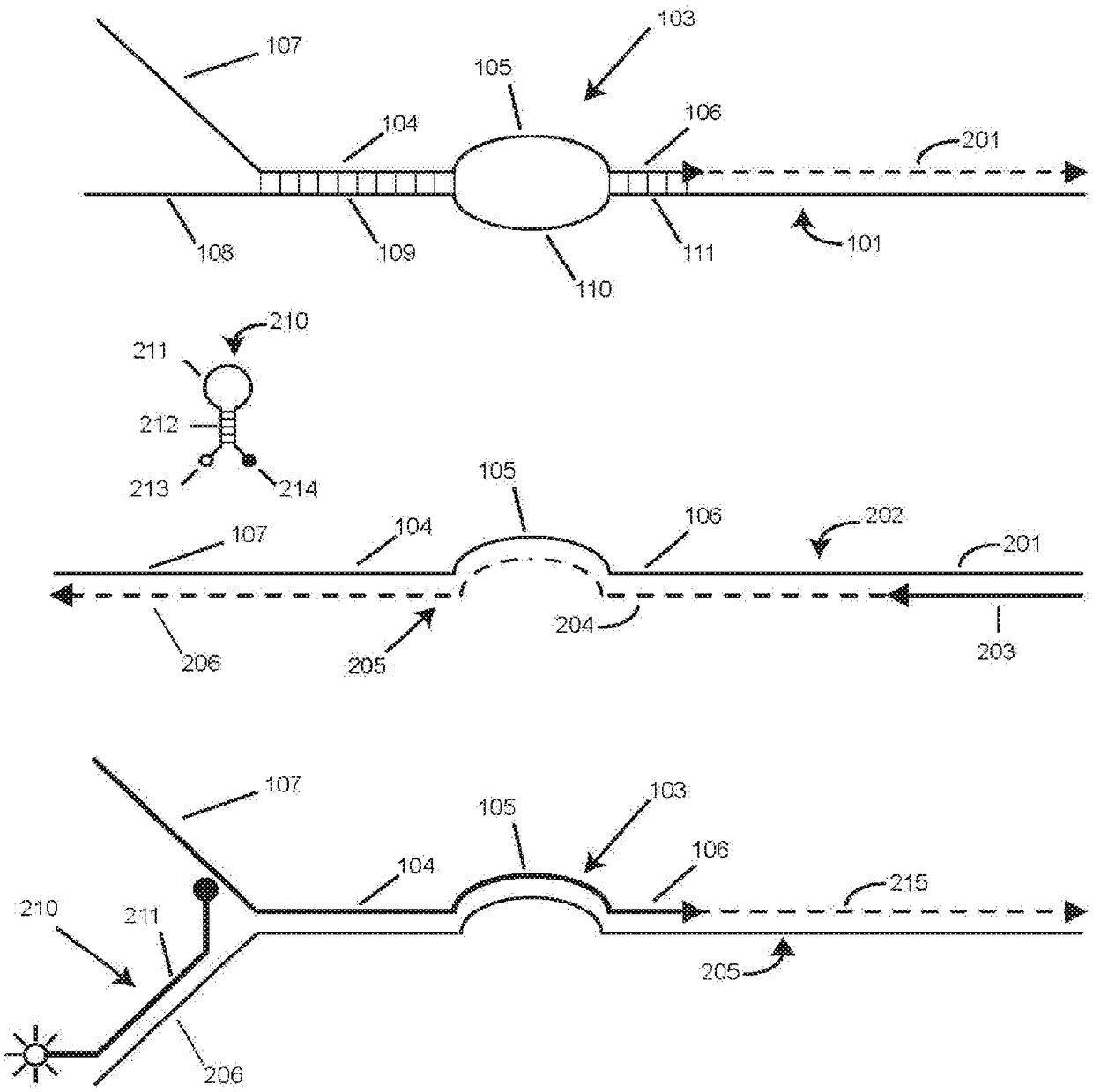
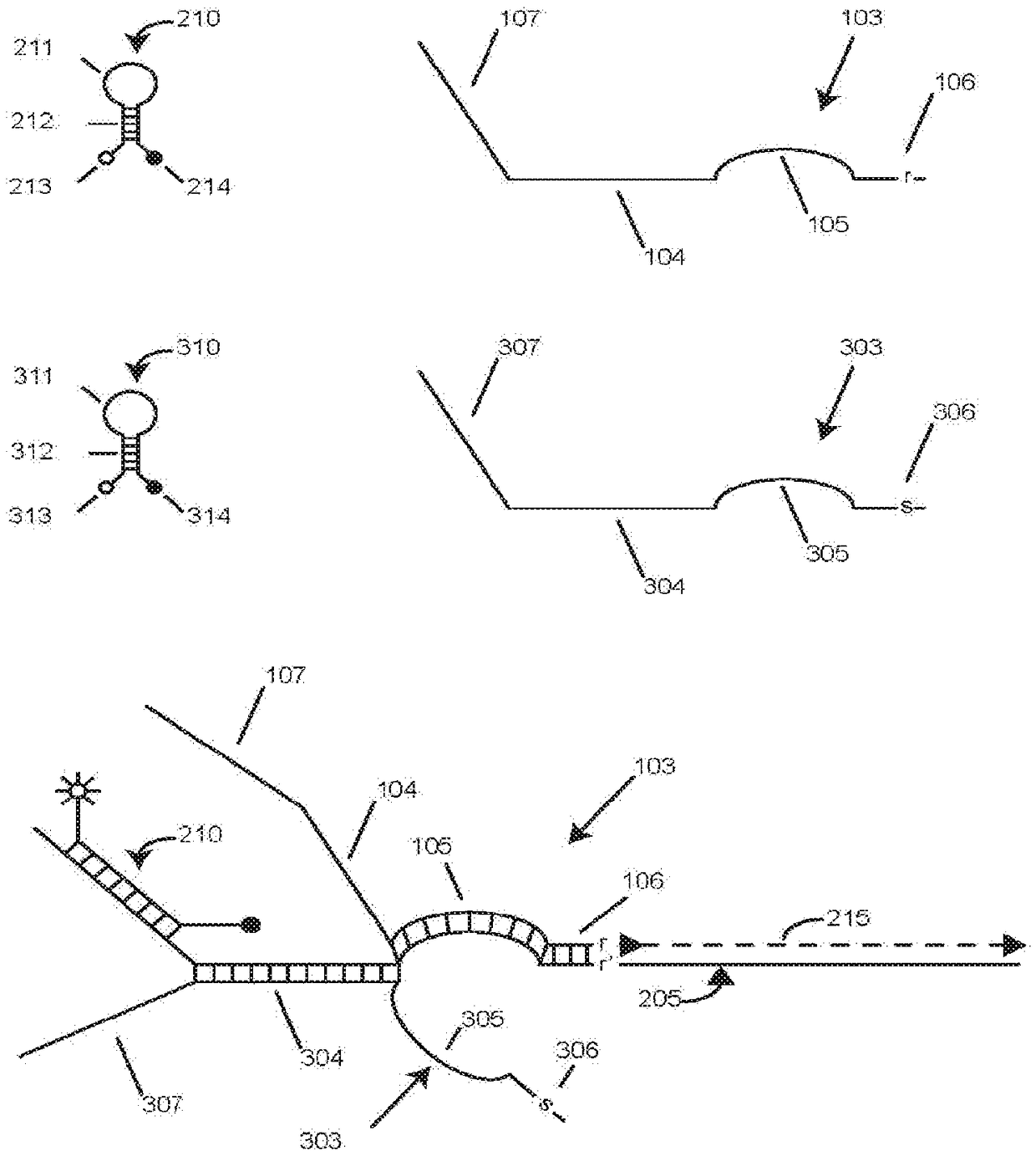
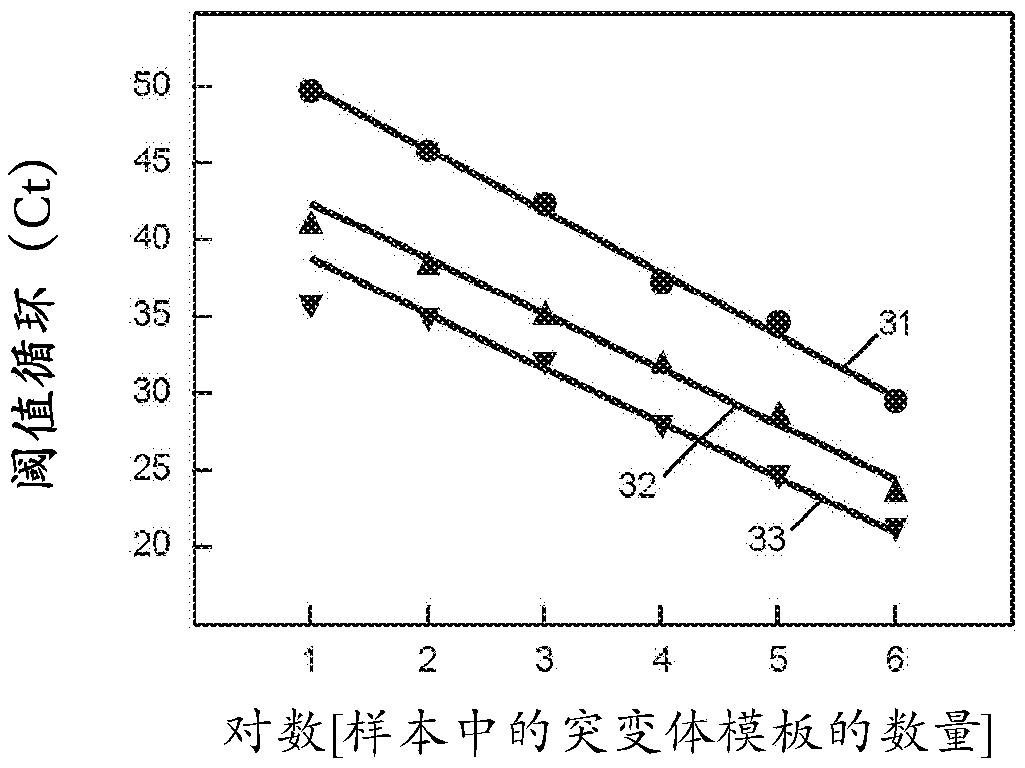
[0090] Example 1 presents a singleplex experiment using restriction enzyme digested plasmid DNA to study the effect of varying foot sequence length. We investigated the effect of shortening the foot length of the SuperSelective primer directed against the EGFR L858R mutant sequence to overcome the possibility that the foot will form a hybrid with the G-C rich sequence present in the EGFR wild-type target sequence. We performed three sets of symmetric PCR assays, each using SuperSelective primers with foot sequences 6, 7, or 8 nucleotides in length. In all other respects, the design of the primers was identical: (i) the interrogation nucleotide was located at the 3' penultimate position of each foot; (ii) the anchor sequence was 24 nucleotides long; and (iii) Both the bridge sequence and the intervening sequence are 14 nucleotides long. Each set of PCR assays used different amounts of mutant templates (10 6 、10 5 、10 4 、10 3 、10 2 and 10 1 copies) at 10 6 Primed in the ...
example 2
[0091] Example 2 presents a singleplex experiment using restriction enzyme digested plasmid DNA to study the effect of varying the perimeter of the vesicle (the length of the bridge sequence plus the length of the intervening sequence plus 4). A single-stranded vesicle functionally separates the anchor hybrid from the foot hybrid (see figure 1 ). We performed three sets of symmetric PCR assays identical to the experiment described in Example 1, except that three different SuperSelective primers were used, each of which when hybridized to its template Form symmetrical bubbles. Each of these three primers has the same 7-nucleotide long foot sequence, where the interrogation nucleotide is located at the 3' penultimate position, and the length of the anchor sequence in each primer is maintained at 24 nucleotides. However, the length of the bridge sequence in each primer was chosen to be 10, 14 or 18 nucleotides, and the identity of the anchor sequence in each primer was chosen ...
example 3
[0092] Example 3 presents a singleplex experiment using restriction enzyme digested plasmid DNA to study the effect of altering the position of a single interrogation nucleotide within the foot sequence, including the ability of the primers to discriminate between mutant and wild-type templates. We performed a series of symmetric PCR assays using six different SuperSelective primers, each with an interrogating nucleotide at a different position within the 7 nucleotide long foot sequence. The lengths of the anchor, bridge and intervening sequences were maintained at 24, 14 and 14 nucleotides, respectively. These are 6:1:0, 5:1:1, 4:1:2, 3:1:3, 2:1:4 and 1:1:5. Two reactions were performed with each primer, one with 10 6 copies of the mutant template were primed, and one was primed with 10 6 copies of the wild-type template are primed. The observed threshold cycles are listed in Table 4 of Example 3. The results show that the threshold cycle (C T ) and wild-type threshold c...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap