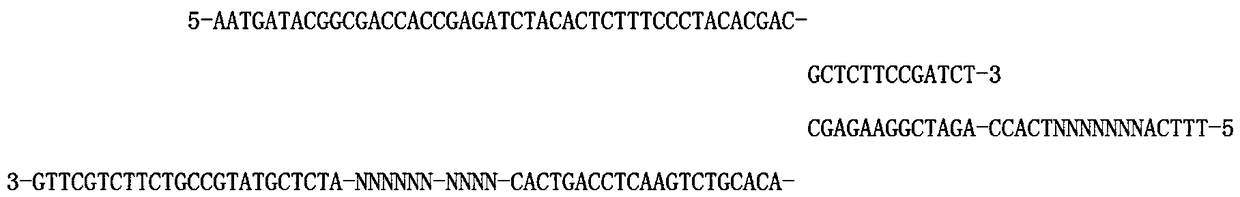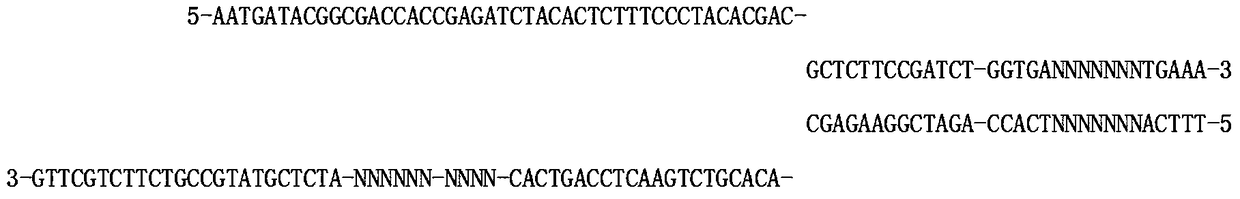Method for preparing new-generation sequencing connector with double chain molecular label
A technology for double-stranded molecules and sequencing joints, which is applied in the field of molecular biology and can solve problems such as the lack of a better method for adding double-stranded tags
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0032] The technical solution of the present invention will be described in detail below through specific embodiments.
[0033] Technical solutions:
[0034] Step 1: Synthesize the linker sequence. Synthesize the following two sequences:
[0035] Sequence 1:
[0036] 5-A*A*TGATACGGCGACCACCGAGATTCTACACTCTTTCCCTACACGAC-GCTCTTCCGAT*C*T-3
[0037] Sequence 2:
[0038] 5-NNNN-ANNNNNNNTCACC-AG*A*TCGGAAGAGC-ACACGTCTGAACTCCAGTCAC-NNNN-NNNNNNNN-ATCTCGTATGCCGTCTTCTGCT*T*G-3
[0039] "*" in the sequence is a protective modification;
[0040] The 1st to 4th bases are the protection bases of restriction endonucleases, the number of bases varies from 0 to 10, and the specific bases are variable, depending on the actual case design;
[0041] The bases from the 6th to the 12th are double-strand molecular tags of 7 random bases;
[0042] The 52nd to 55th bases are single-strand molecular tags with 4 random bases, and the number of bases can be changed according to the application;
[0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com