Intracellular scaffold, plasmids constructed by intracellular scaffold and application of plasmids
A technology of internal scaffolds and cells, which is applied in the direction of antibody mimics/scaffolds, microbial-based methods, and the use of vectors to introduce foreign genetic materials, etc. It can solve the problems of easy degradation, limited ligand protein types, scalability and stability bottlenecks, etc. Problems, to achieve the effect that is conducive to the aggregation of enzyme proteins and strong copolymerization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
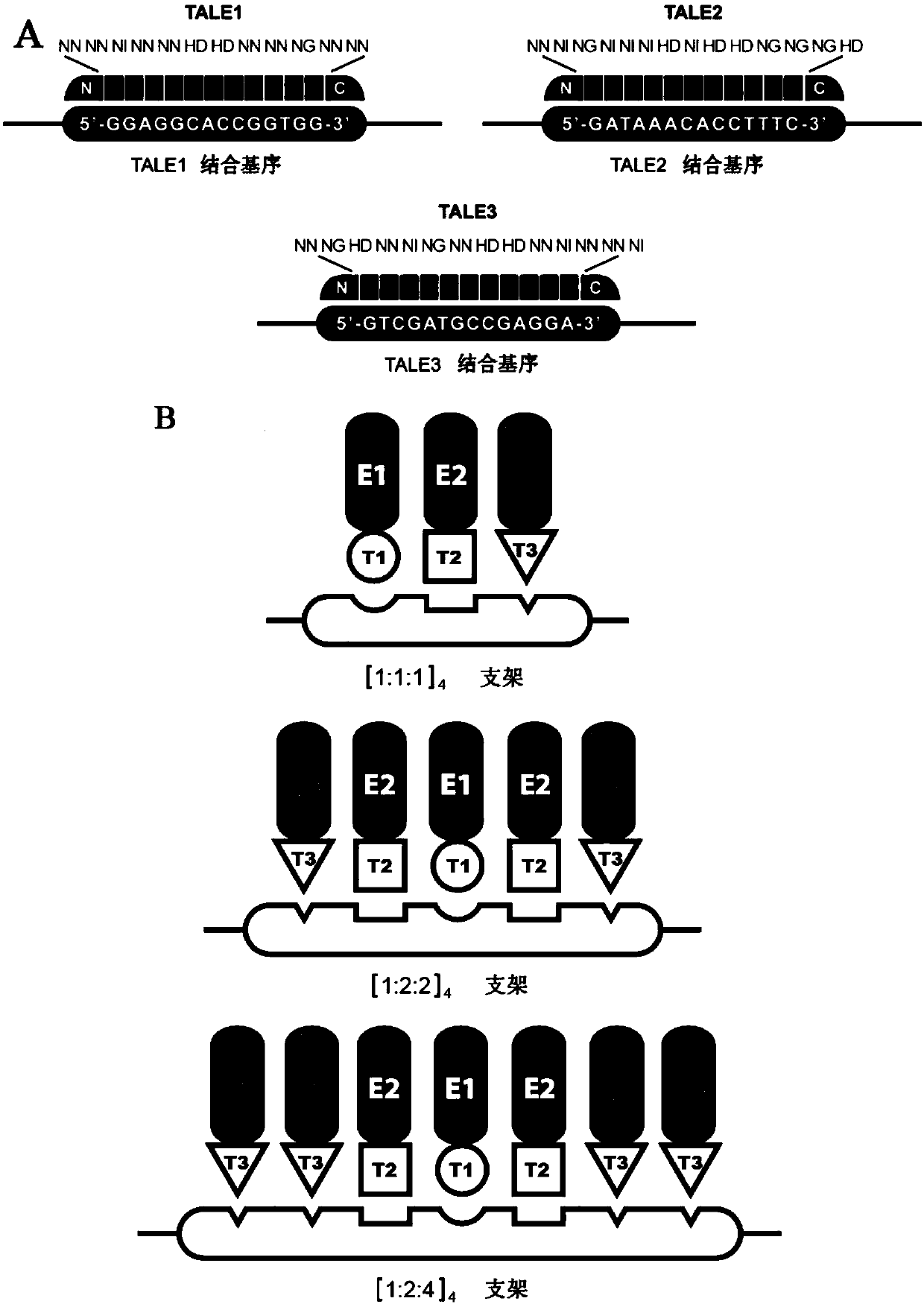
[0080] A TALE-DNA cell scaffold comprises DNA scaffold sequence SCAF1 and TALE-target molecule fusion protein.
[0081] The DNA scaffold sequence SCAF1 includes the binding sequence BM1, the binding sequence BM2 and the binding sequence BM3; wherein, BM1, BM2, and BM3 are concatenated according to the copy number of 1:1:1, and a 6bp length is designed between BM1, BM2, and BM3. interval sequence. BM1, BM2, and BM3 are arranged in a 1:1:1 manner to form a scaffold structure unit repeated four times.
[0082] BM1 (SEQ ID NO.1): 5'-ggaggcaccggtgg-3';
[0083] BM2 (SEQ ID NO.2): 5'-gataaacacctttc-3';
[0084] BM3 (SEQ ID NO. 3): 5'-gtcgatgccgagga-3'.
[0085] 6bp spacer sequence (SEQ ID NO.7): 5'-acagtt-3'
[0086] The DNA scaffold sequence SCAF1 is (SEQ ID NO.9):
[0087] ggaggcaccggtgg acagtt gataaacacctttc acagtt gtcgatgccgagga acagtt ggaggcac cggtgg acagttg ataaacacctttc acagtt gtcgatgccgagga acagtt ggaggcaccggtgg acagtt gata aacacctttc acagtt gtcgat...
Embodiment 2
[0102] A TALE-DNA cell scaffold, comprising DNA scaffold sequences SCAF1, SCAF2, SCAF3, SCAF4, SCAF5, SCAF6 and TALE1 / 2 / 3-target molecule 1 / 2 / 3 fusion protein.
[0103] The DNA scaffold sequence SCAF1 is consistent with Example 1.
[0104] The DNA scaffold sequence SCAF2, BM1, BM2 and BM3 were concatenated according to the copy number ratio of 1:1:1, and a spacer sequence with a length of 16 bp was designed between BM1, BM2 and BM3. BM1, BM2, and BM3 were arranged in a 1:1:1 manner to form a scaffold structure unit repeated four times.
[0105] The 16bp spacer sequence is: 5'-acagtagctacaacct-3' (SEQ ID NO.8).
[0106] The DNA scaffold sequence SCAF2 is (SEQ ID NO.10):
[0107] ggaggcaccggtgg acagtagctacaacct gataaacacctttc acagtagctacaacct gtcgatgc cgagga acagtagctacaacct ggaggcaccggtgga cagtagctacaacct gataaacacctttc acagtagctacaacct gtcgatgccgagga acagtagctacaacct ggaggcaccggtgg acagtagctacaacct gataaacacc tttc acagtagctacaacct gtcgatgccgagga acagtag...
Embodiment 3
[0129] A TALE-DNA cell scaffold, comprising DNA scaffold sequences SCAF1, SCAF2, SCAF3, SCAF4, SCAF5, SCAF6 and TALE1 / 2 / 3-target molecule A / B / C fusion protein. .
[0130] The DNA scaffold sequences SCAF1, SCAF2, SCAF3, SCAF4, SCAF5, SCAF6 are the same as in Example 1.
[0131] Among the TALE-target molecule fusion proteins, the target molecules are AtoB, HMGS, and HMGR; they are fused with TALE1, TALE2, and TALE3 to form TALE1-AtoB fusion protein, TALE2-HMGS fusion protein, and TALE3-HMGR fusion protein. The fusion method is consistent with Example 1.
[0132] TALE1-AtoB fusion protein specifically binds to the binding sequence BM1; TALE2-HMGS fusion protein specifically binds to the binding sequence BM2; TALE3-HMGR specifically binds to the binding sequence BM3.
[0133] The TALE expression sequence was constructed using Golden Gate TALEN and TAL Effector Kit 2.0, and the DNA scaffold sequence was synthesized by DNA synthesis. Utilizing the properties of restriction endonu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com