The invention discloses a data classification method for single cell sequencing
A technology for single-cell sequencing and data classification, applied in sequence analysis, instruments, calculations, etc., can solve problems such as lack of methods, and achieve fast and efficient classification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
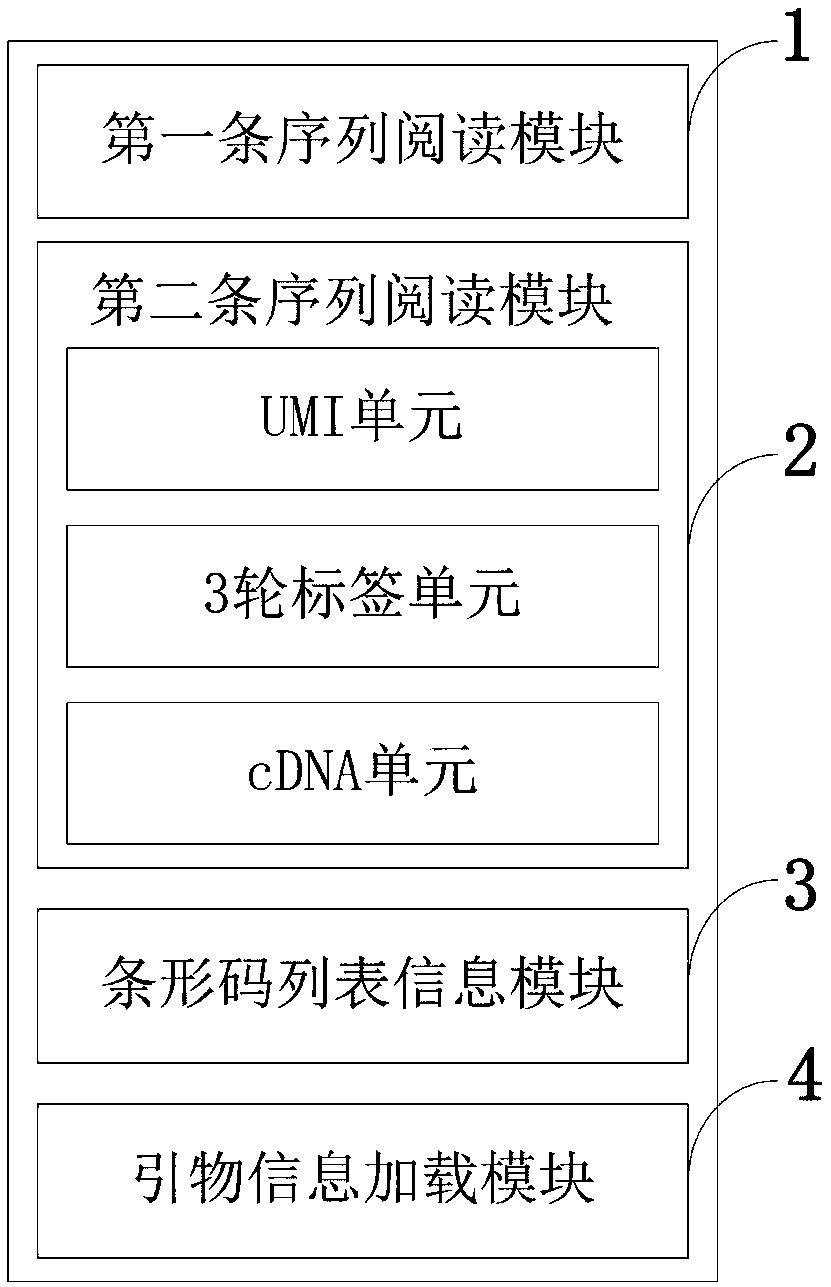
[0067] 1. Classification and extraction of Read2.fastq data
[0068] Such as image 3 As shown, it is the data content of Read2. Read2 is divided into 5 parts, UMI, 3 rounds of tags and cDNA, where UMI and 3 rounds of tags are used as identifiers to classify different cell sources, and cDNA is the final sequence information to be extracted .
[0069] (1) Firstly extract 3 rounds of barcode from the sequence. The specific method is to first find out the position of the characteristic sequence in the sequence, and then shift forward 8 bits to extract the corresponding barcode. When searching for the position of the characteristic sequence, the K-mer method is adopted, and a fault-tolerant mechanism is provided. After extracting 3 rounds of barcode, convert the barcode into 3 sets of numbers through the Barcode Table, and use them together as a unique identifier to determine a cell. Then append the UMI to the identity.
[0070] After the feature sequence is obtained, the barc...
Embodiment 2
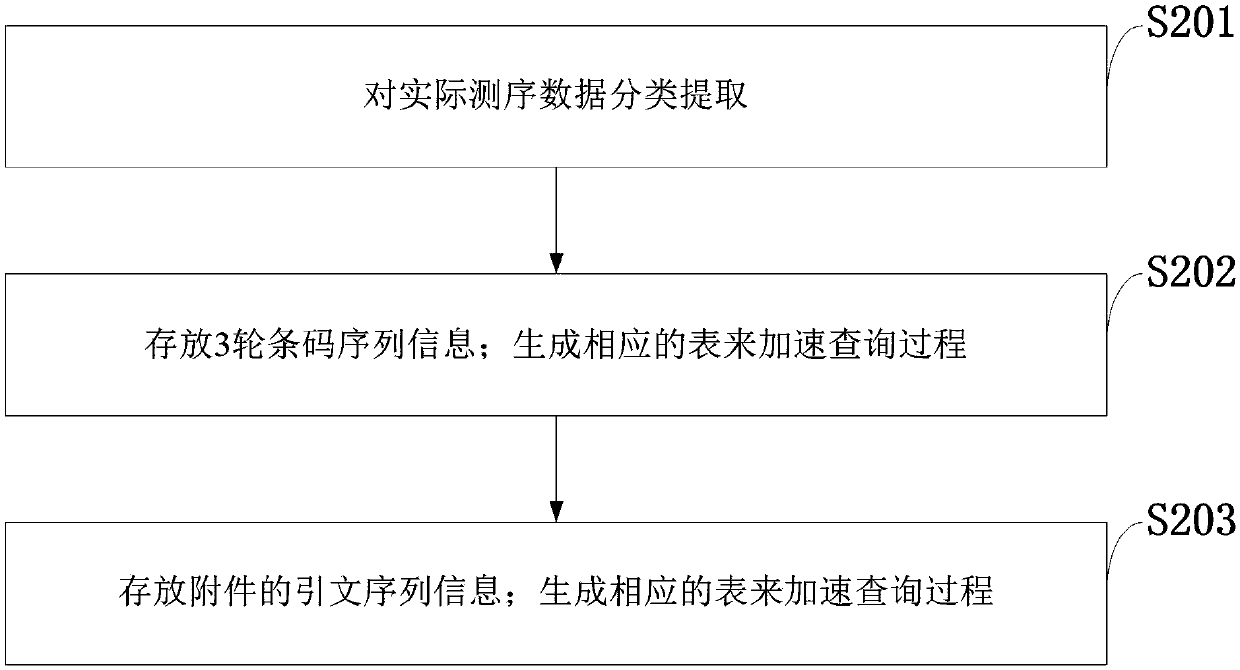
[0103] Step 1, load the actual data and related files:
[0104] 2 actual data files:
[0105] R1.fastq
[0106] R2.fastq
[0107] Three rounds of barcode information files:
[0108]BarcodeList
[0109] Feature information:
[0110] PrimerList
[0111] Step 2: Generate corresponding tables according to BarcodeList and PrimerList to speed up the query process: generate 3 tables according to the three rounds of information of BarcodeList, such as Image 6 :
[0112] Generate PrimerTable based on PrimerList such as Figure 7 :
[0113] The generation of PrimerTable is a linked list array generated according to the input text file PrimerList. Treat the data in PrimerList as a whole long sequence, take a fragment of length k each time, start to take the fragment from the beginning, and shift backward by 1 bit each time, mainly because the record subsequence appears in the whole s position.
[0114] Among them, each fragment is converted once, and it is regarded as a 4-ary...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com