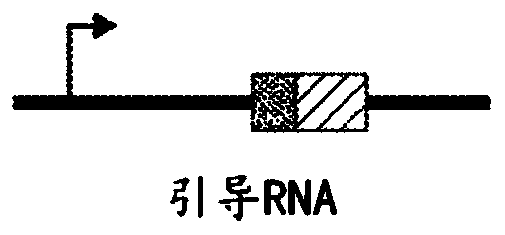
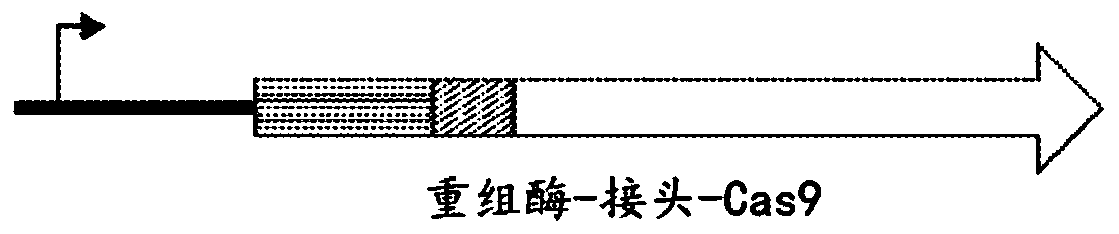
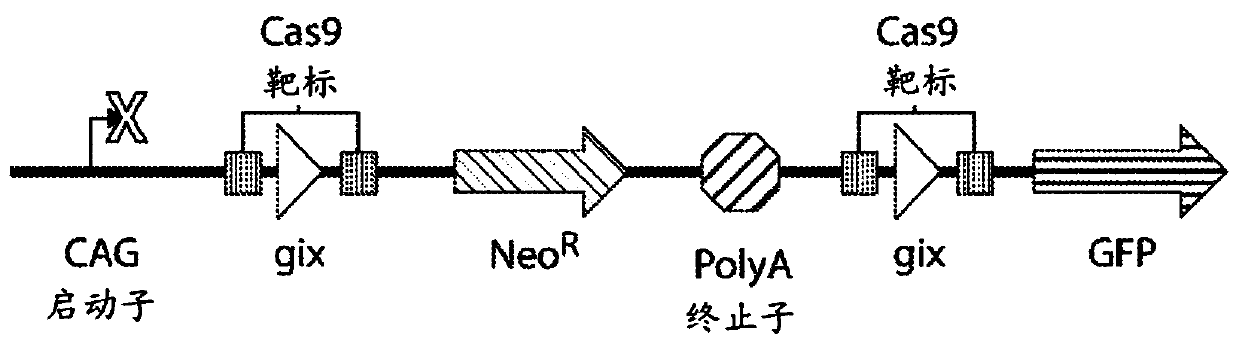
Programmable cas9-recombinase fusion proteins and uses thereof
A fusion protein and recombinase technology, applied in the fields of insertion, inversion, and deletion, which can solve problems such as limited applications
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0225] Example 1: Programmable Cas9-serine recombinase fusion protein that functions on DNA sequences in mammalian cells
[0226] Materials and methods
[0227] Oligonucleotides and PCR
[0228] All oligonucleotides were purchased from Integrated DNA Technologies (IDT, Coralville, CA) and are listed in Tables 1-5. Enzymes were purchased from New England Biolabs (Ipswich, MA) unless otherwise stated. Plasmid SafeATP-dependent DNase was purchased from Epicenter (Madison, WI). All assembled vectors were transformed into One Shot Mach1-T1 phage-resistant chemically competent cells (Fisher Scientific, Waltham, MA). Unless otherwise stated, all PCR reactions were performed using the Q5 Hot Start High-Fidelity 2X Master Mix. Phusion polymerase is used for circular polymerase extension clone (CPEC) assembly.
[0229] Table 1: Oligonucleotides used for gRNA construction
[0230]
[0231] Table 2: Oligonucleotides and gBlocks used for reporter construction
[0232]
[023...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com