Method for predicting element being rich in riboSnitch or with lost riboSnitch in cancer genome and related equipment
A genomics and cancer technology, applied in genomics, proteomics, instrumentation, etc., can solve problems such as unclear role of riboSNitch
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
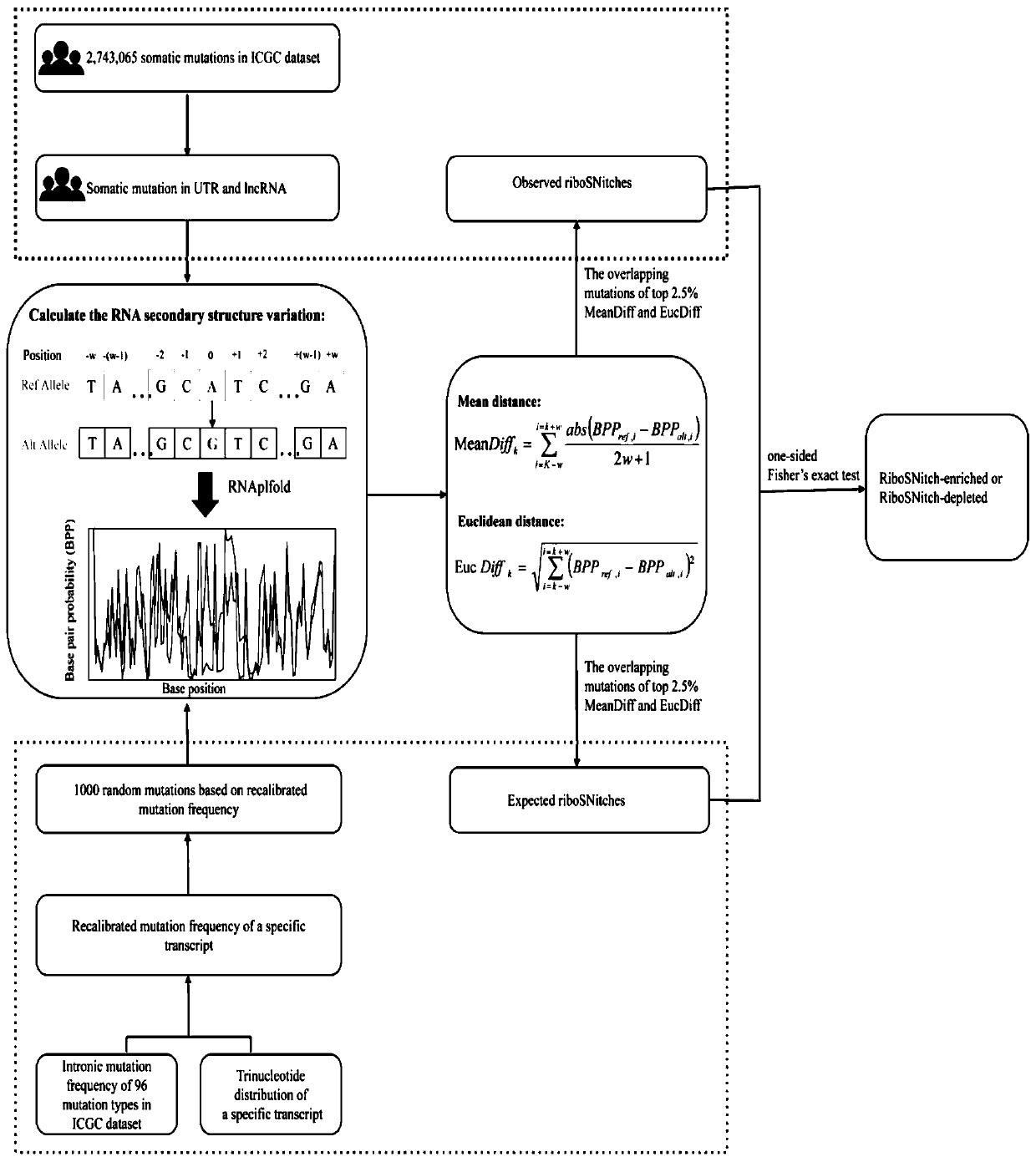
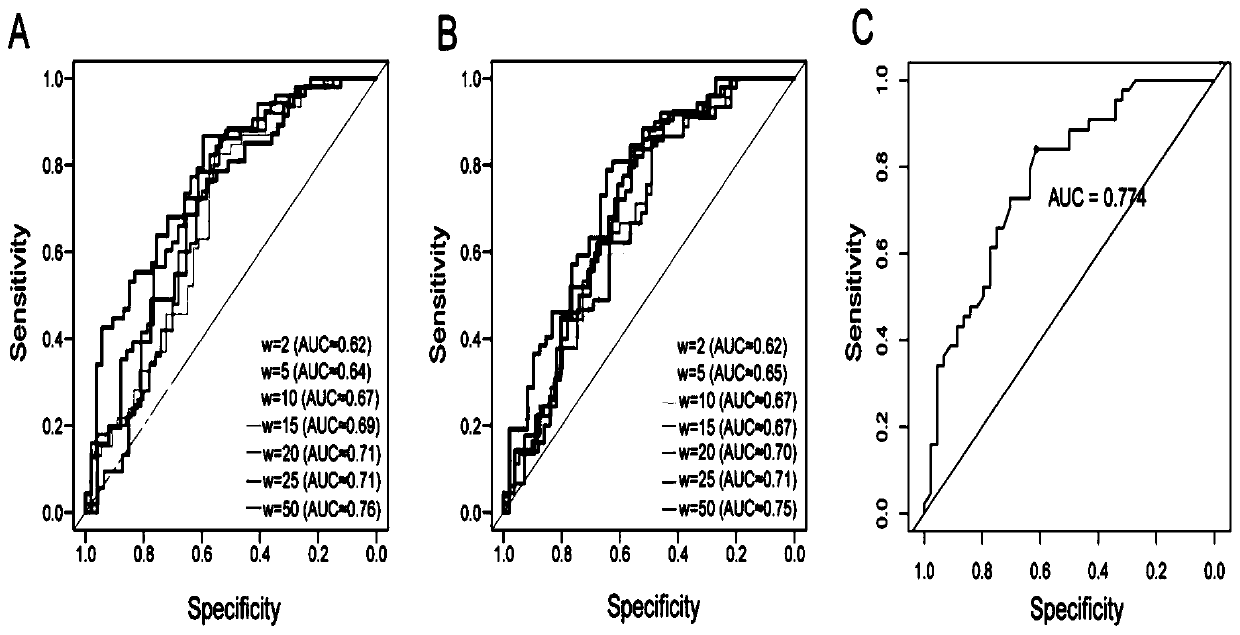
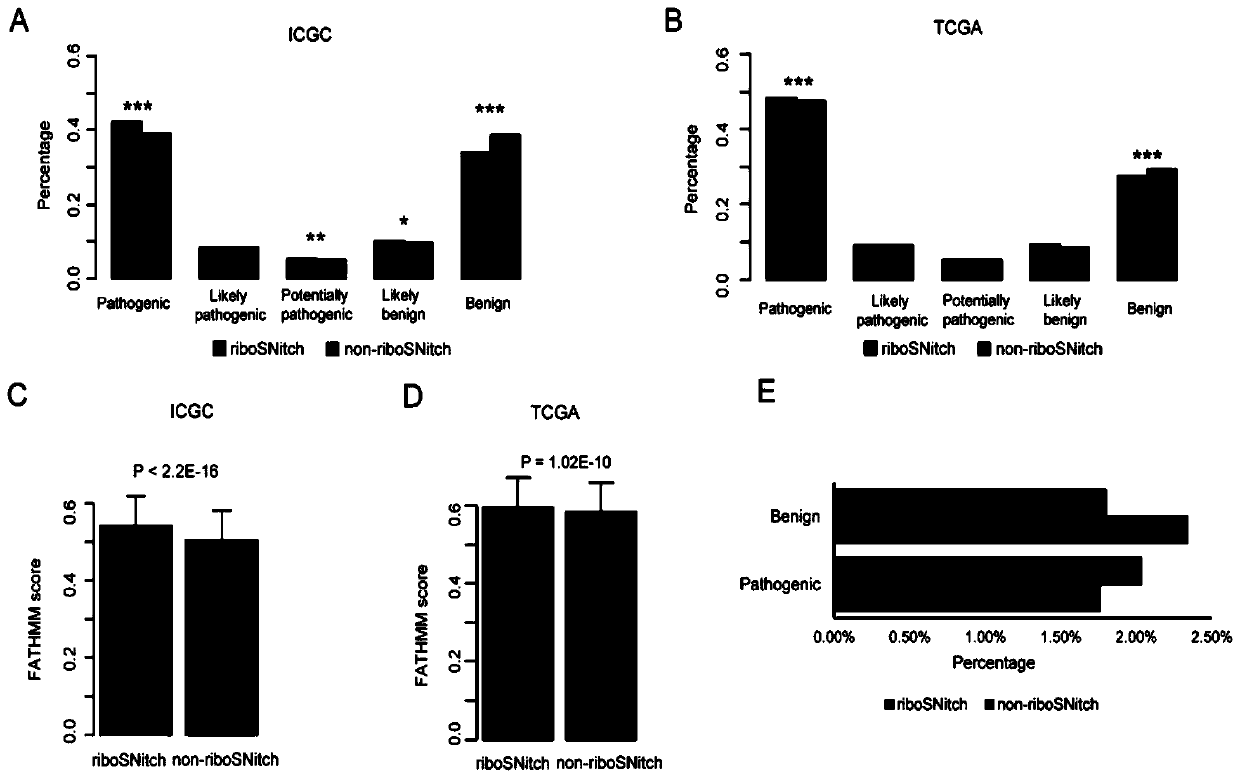
[0049] 1.1 Materials and methods
[0050] 1.1.1 Data collection
[0051] Most cancer somatic mutation data used in this study came from ICGC and TCGA databases. In addition, we collected somatic mutations in 25 whole-genome sequenced melanomas and 100 whole-genome sequenced gastric cancers 20,21 . The mutation data set of normal people is obtained from the Thousand Genomes database, and the present invention uses the data of Phase 3 22 .
[0052] First, we filtered out all indels and only kept point mutations for further analysis. Then, use the UCSC liftOver toolkit to change all hg38-annotated mutations to hg19 mutations23 . In order to remove low-confidence SNVs, we filter the somatic mutations of the cancer database and the germline mutations of the Thousand Genomes. The filtered interval collection is downloaded from the Broad Institute (https: / / personal.broadinstitute.org / anshul / projects / encode / rawdata / blacklists). This list is a collection of all blacklisted regi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com