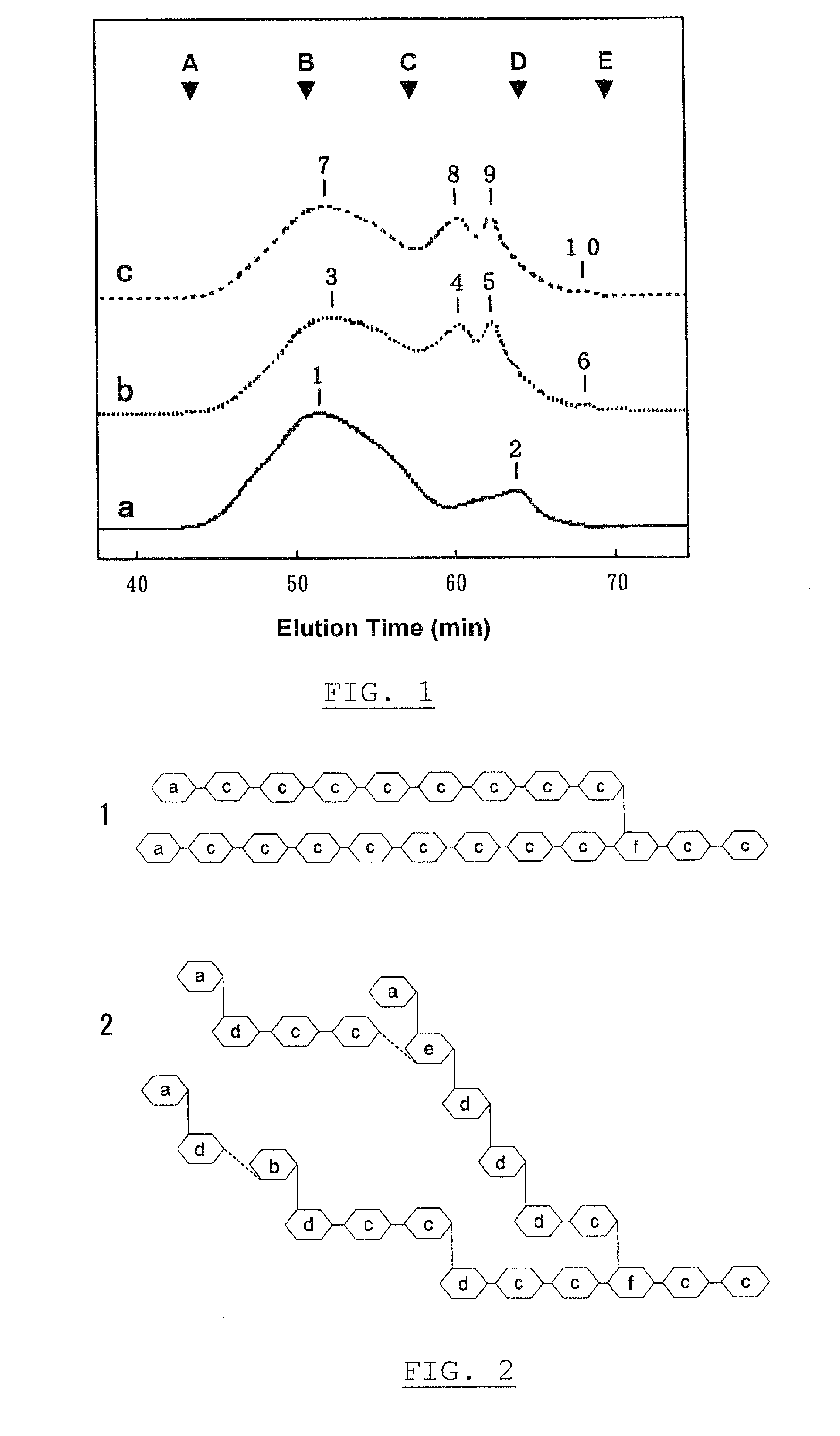
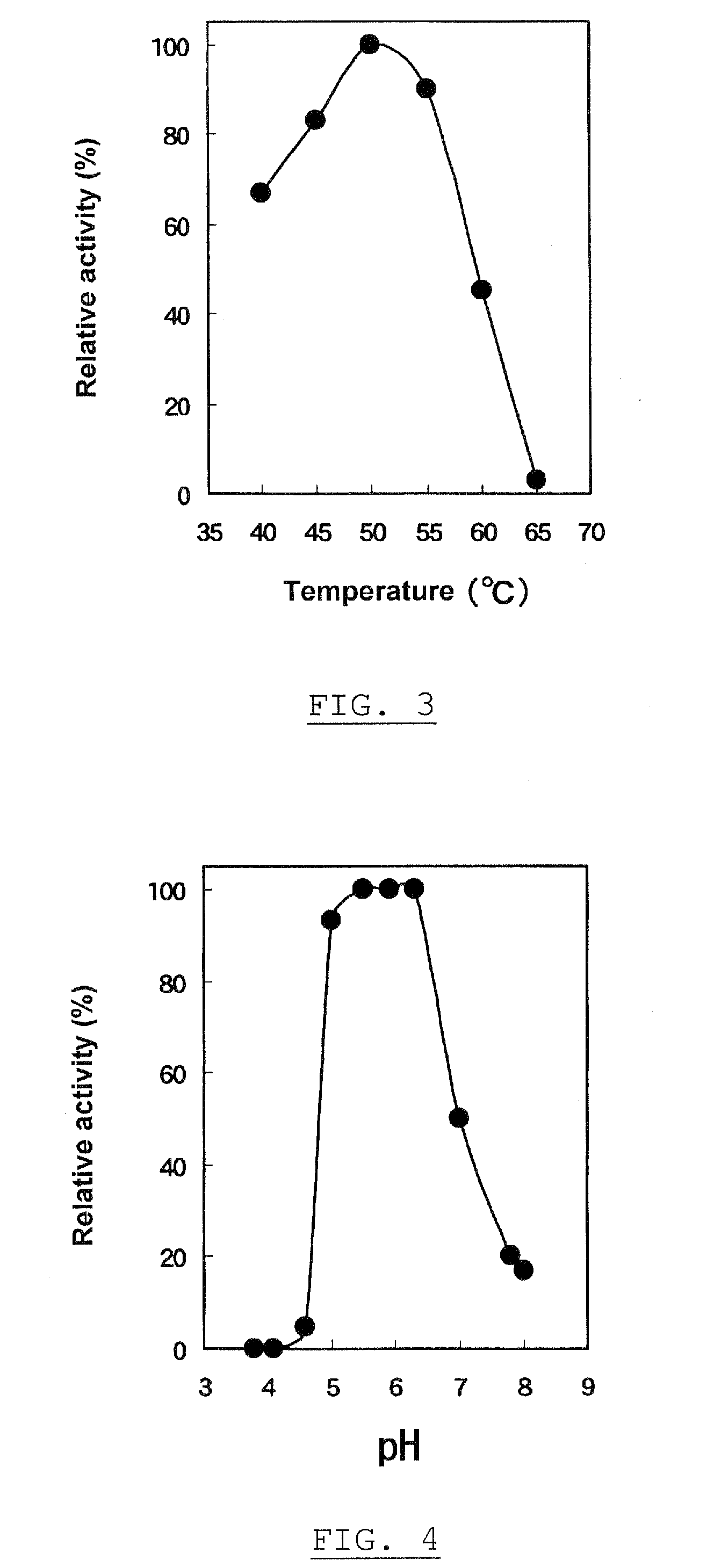
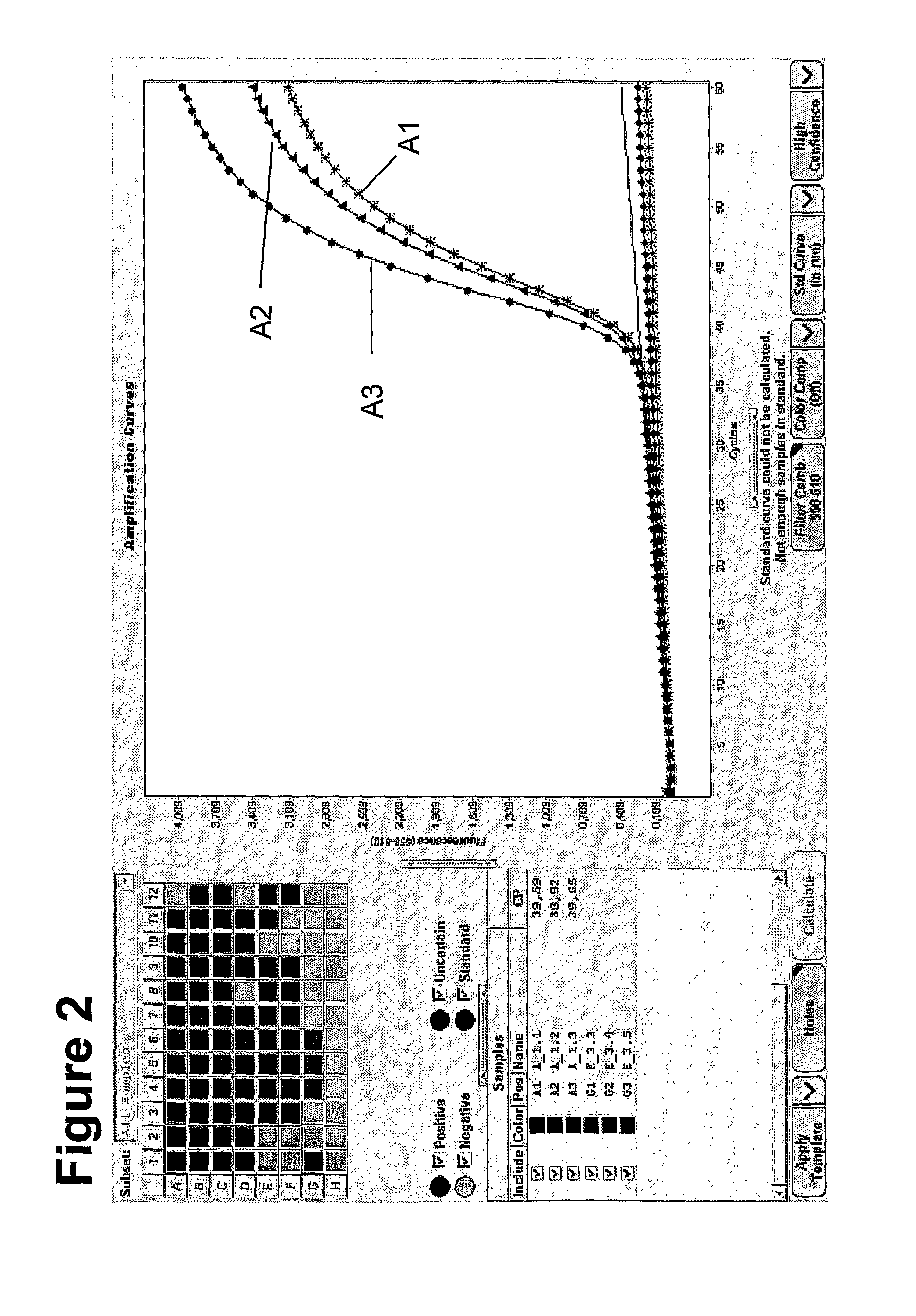
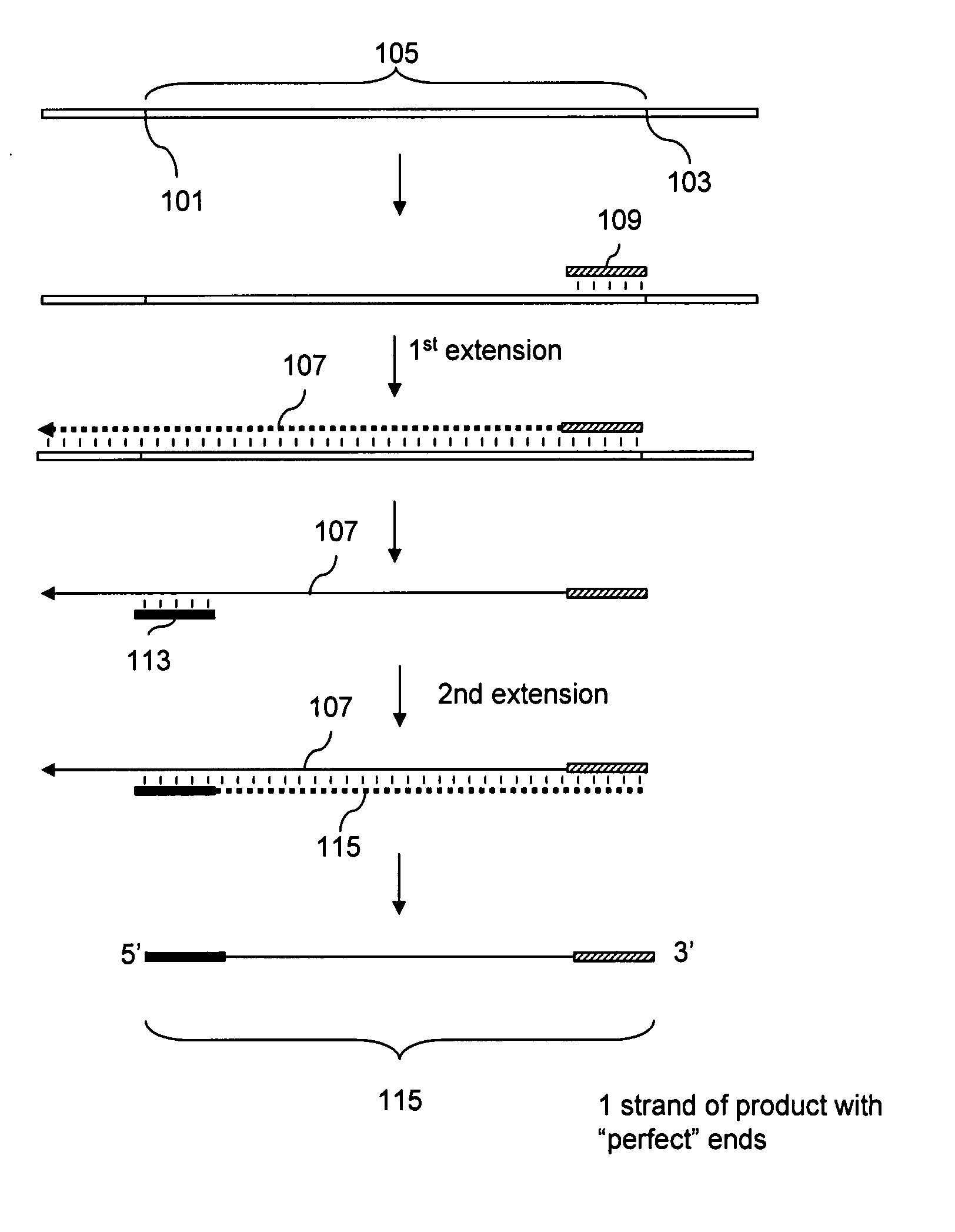
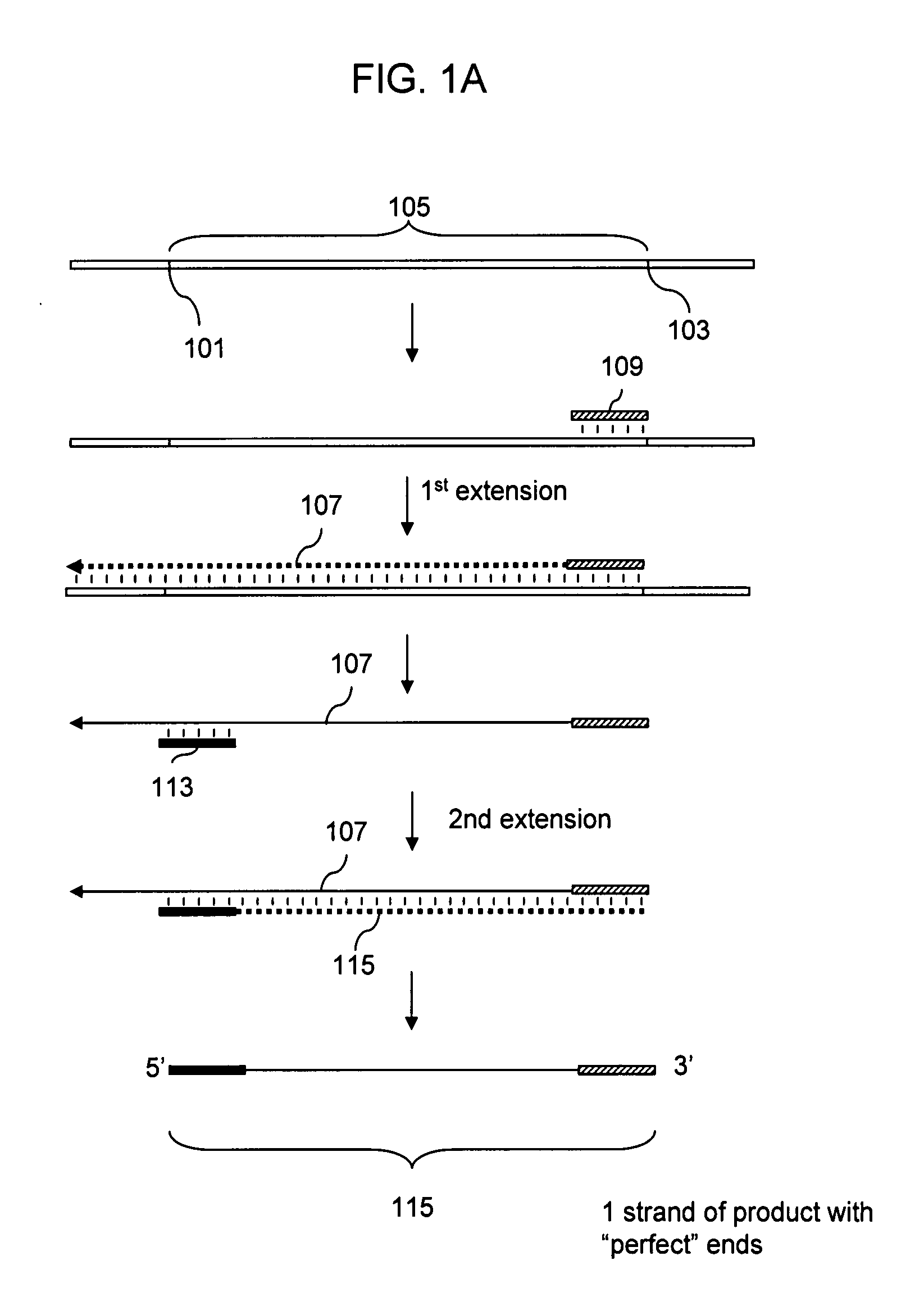
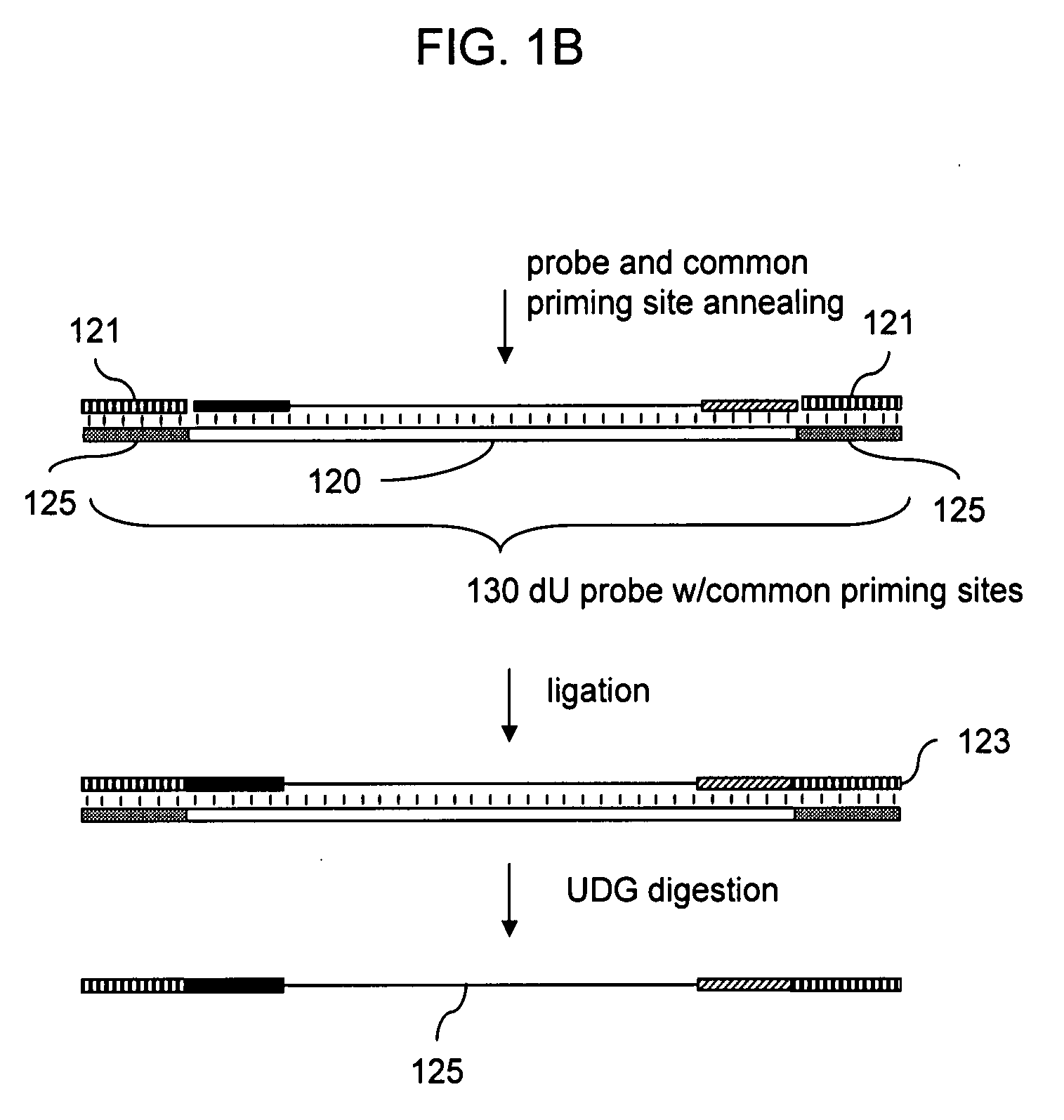
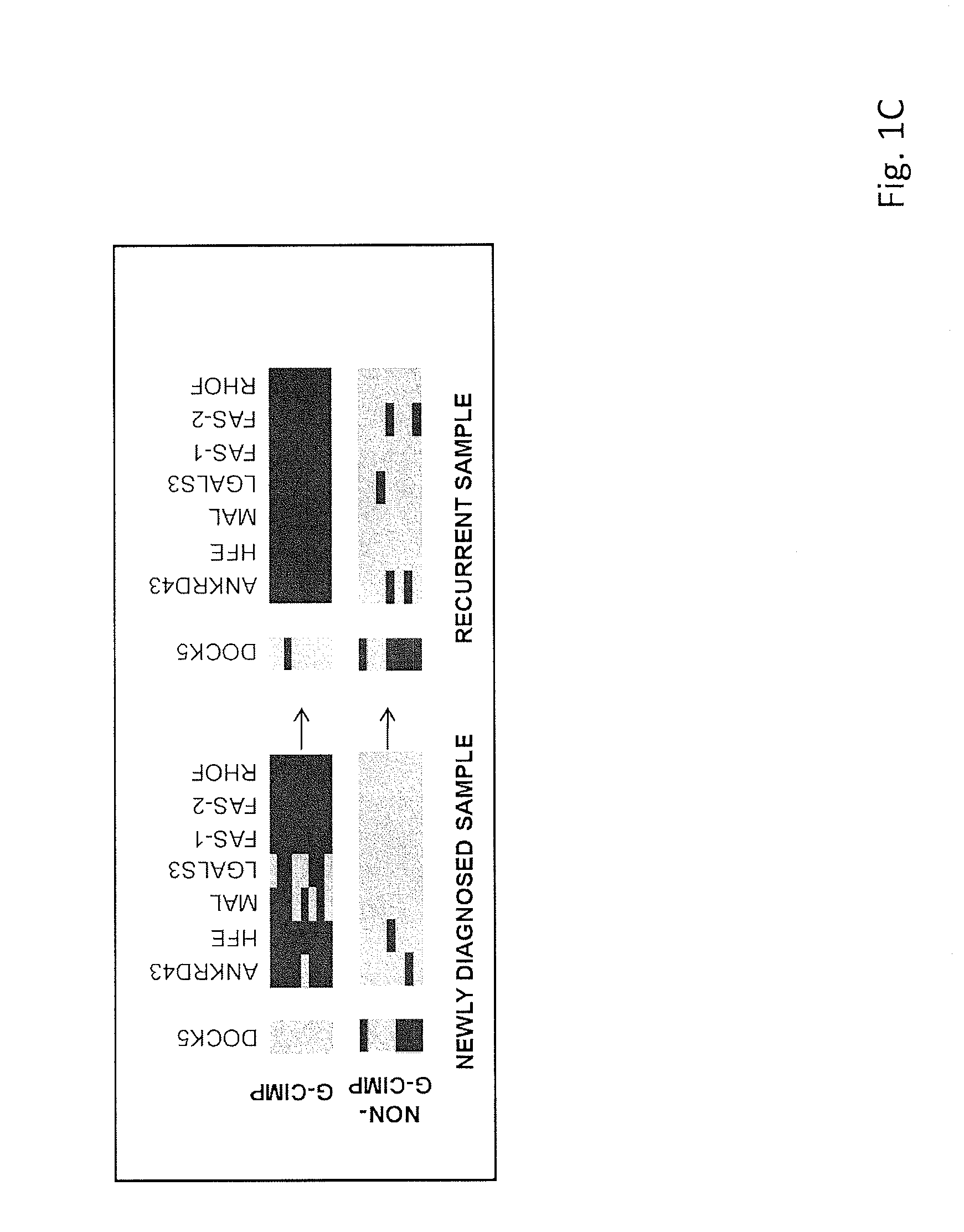
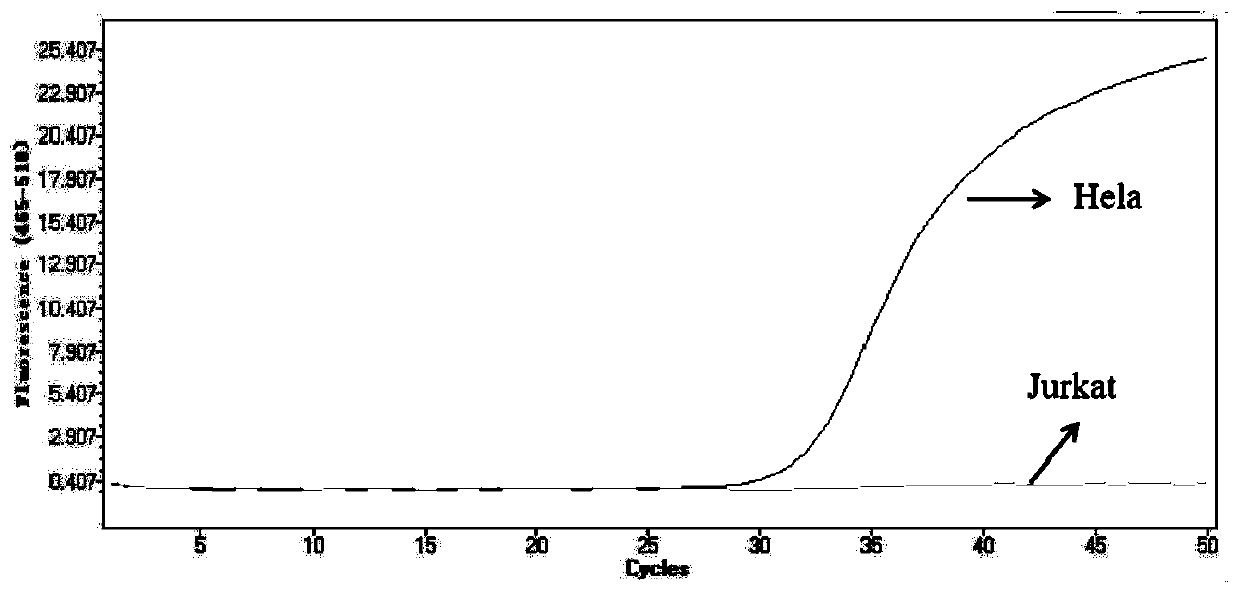
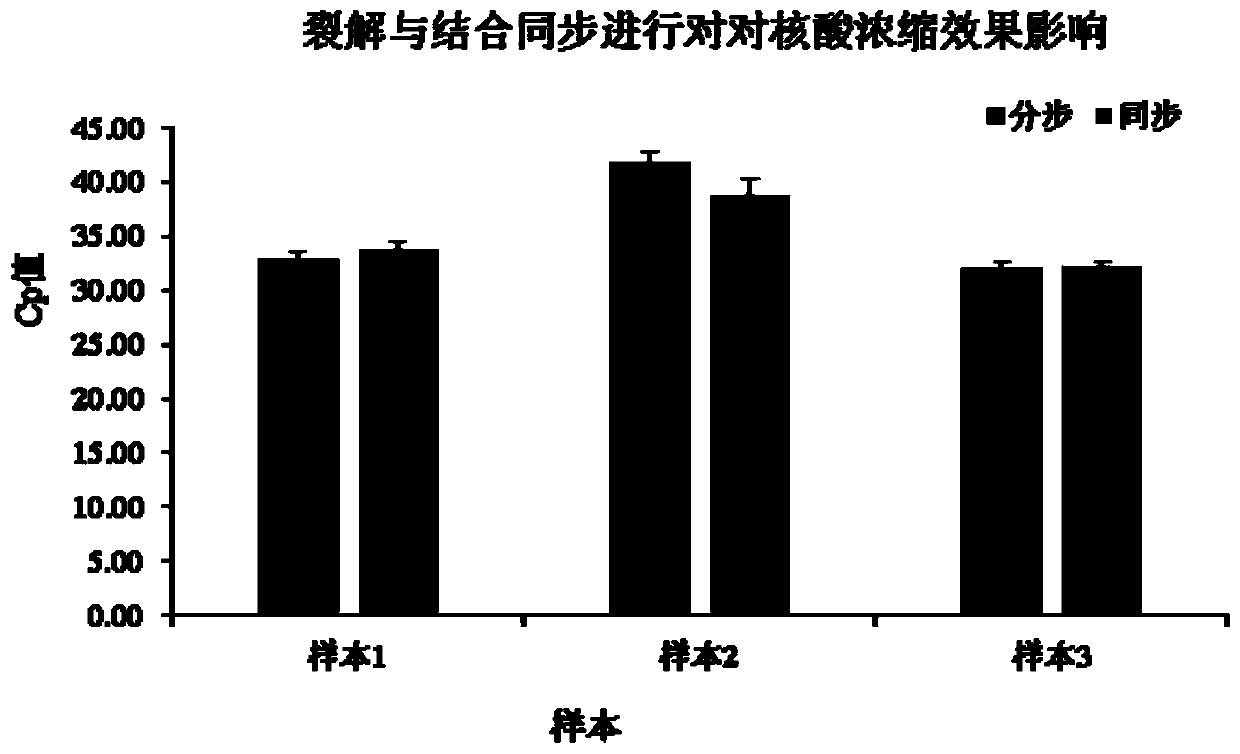
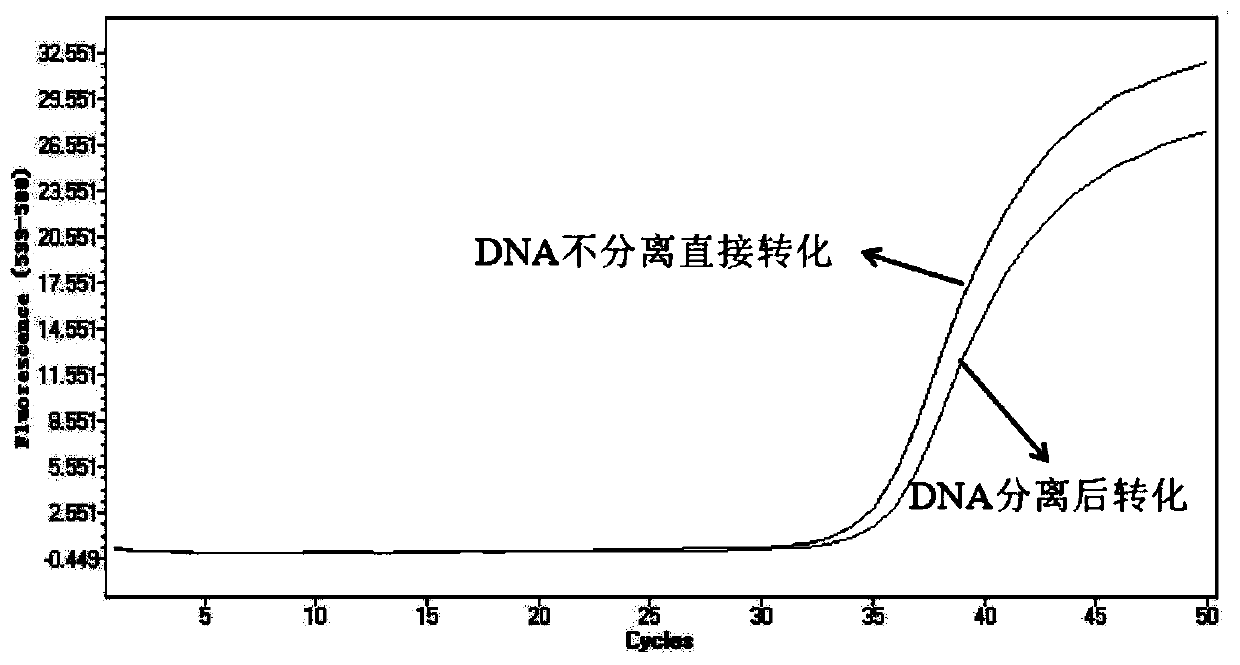
Patents
Literature
72 results about "Methylation analysis" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
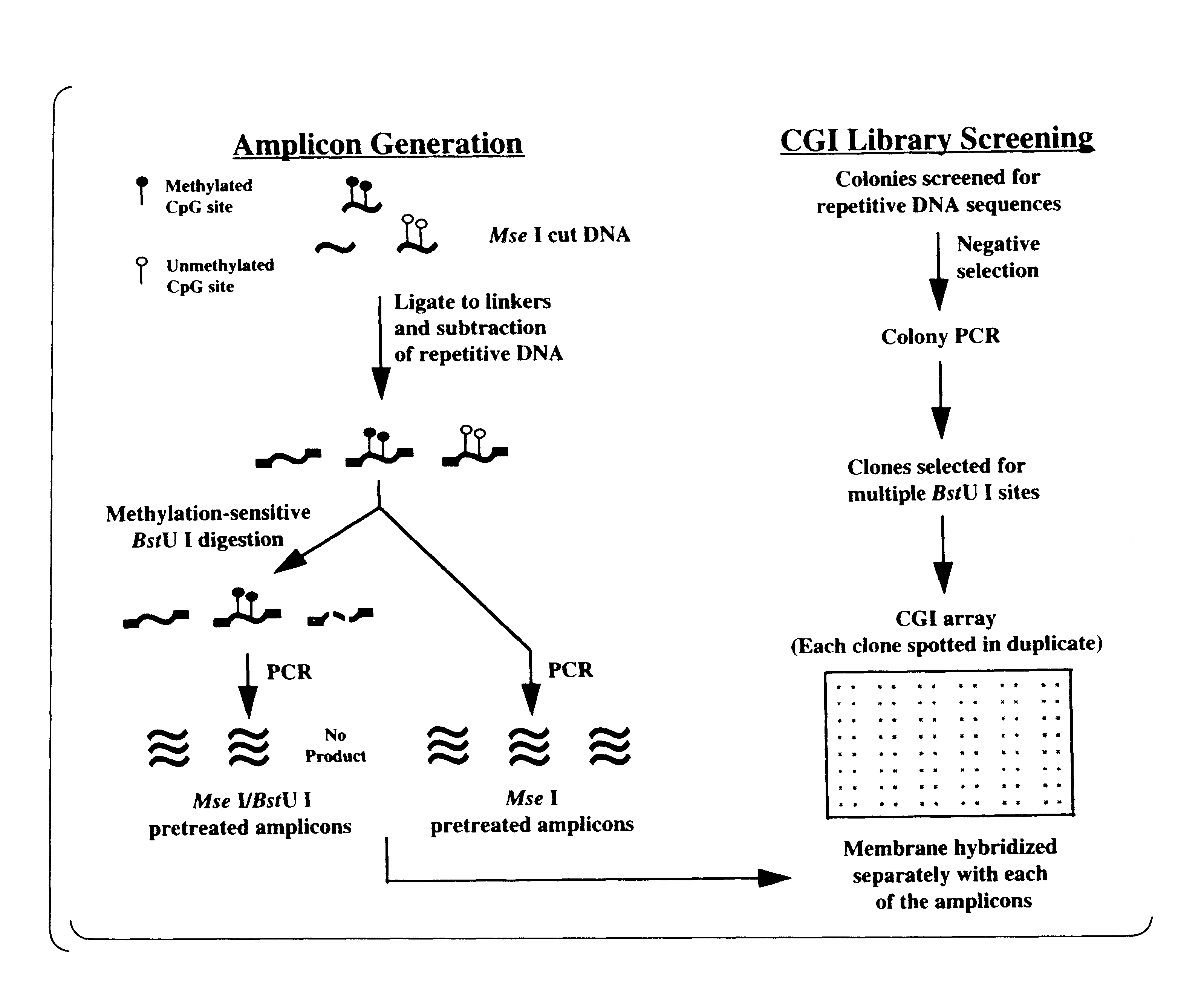
High-throughput methods for detecting DNA methylation
InactiveUS6605432B1Bioreactor/fermenter combinationsBiological substance pretreatmentsDNA methylationHybridization probe
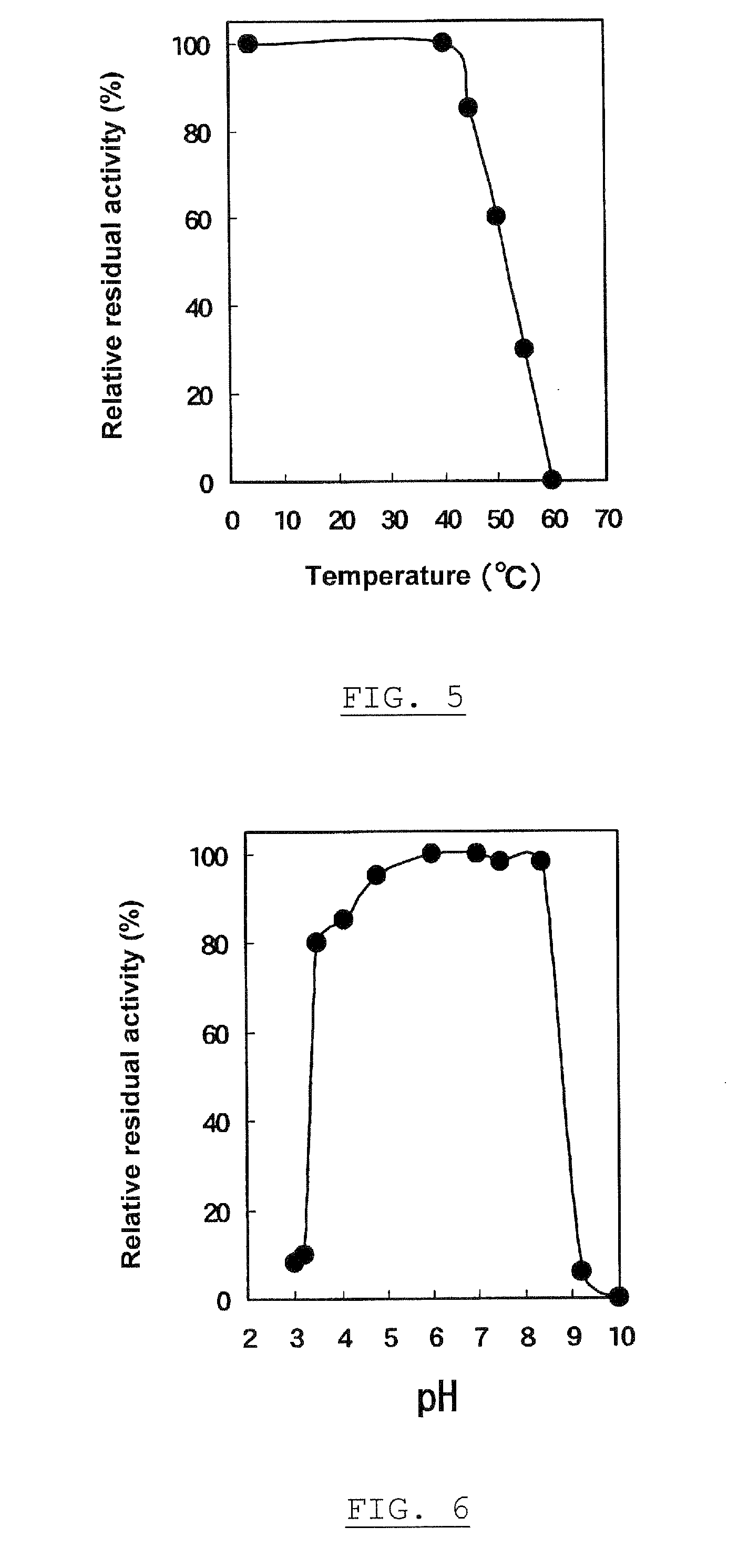
The present invention provides a method of hybridization, differential methylation hybridization (DMH) for high throughput methylation analysis of multiple CpG island loci. DMH utilizes nucleic acid probes prepared from a cell sample to screen numerous CpG dinucleotide rich fragments affixed on a screening array. Positive hybridization signals indicate the presence of methylated sites. Methods of preparing the hybridization probes and screening array are also provided.
Owner:UNIVERSITY OF MISSOURI
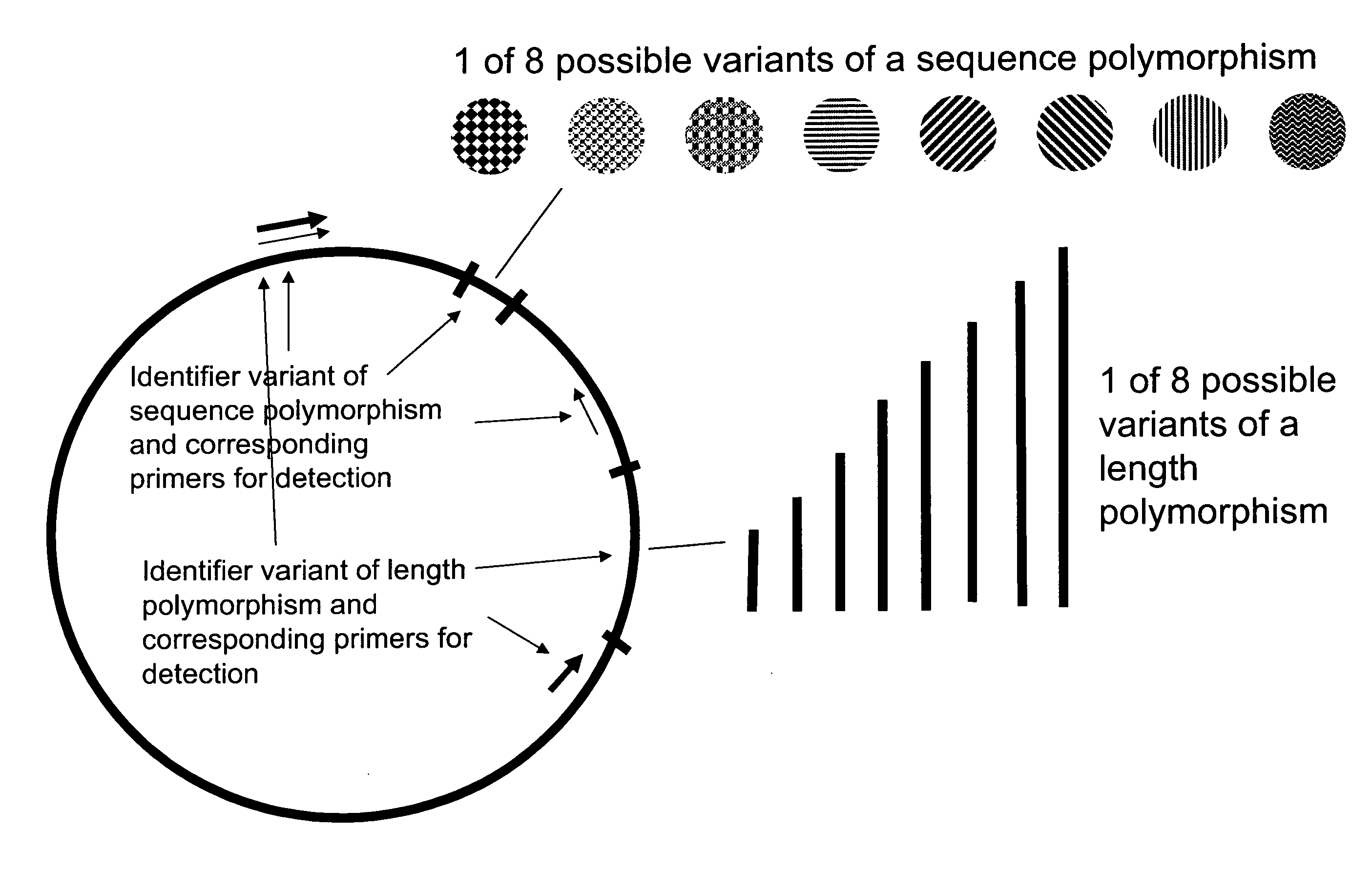
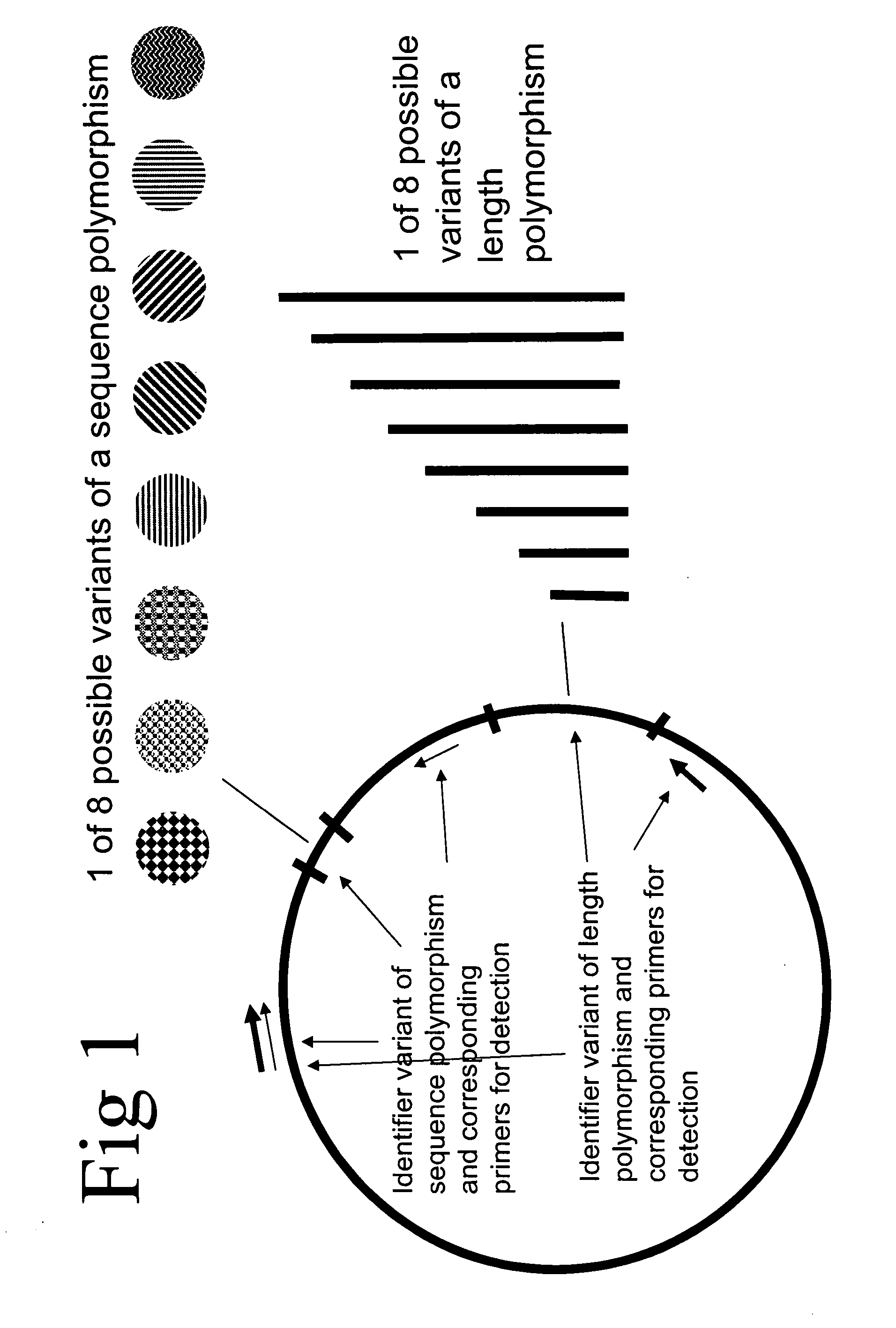
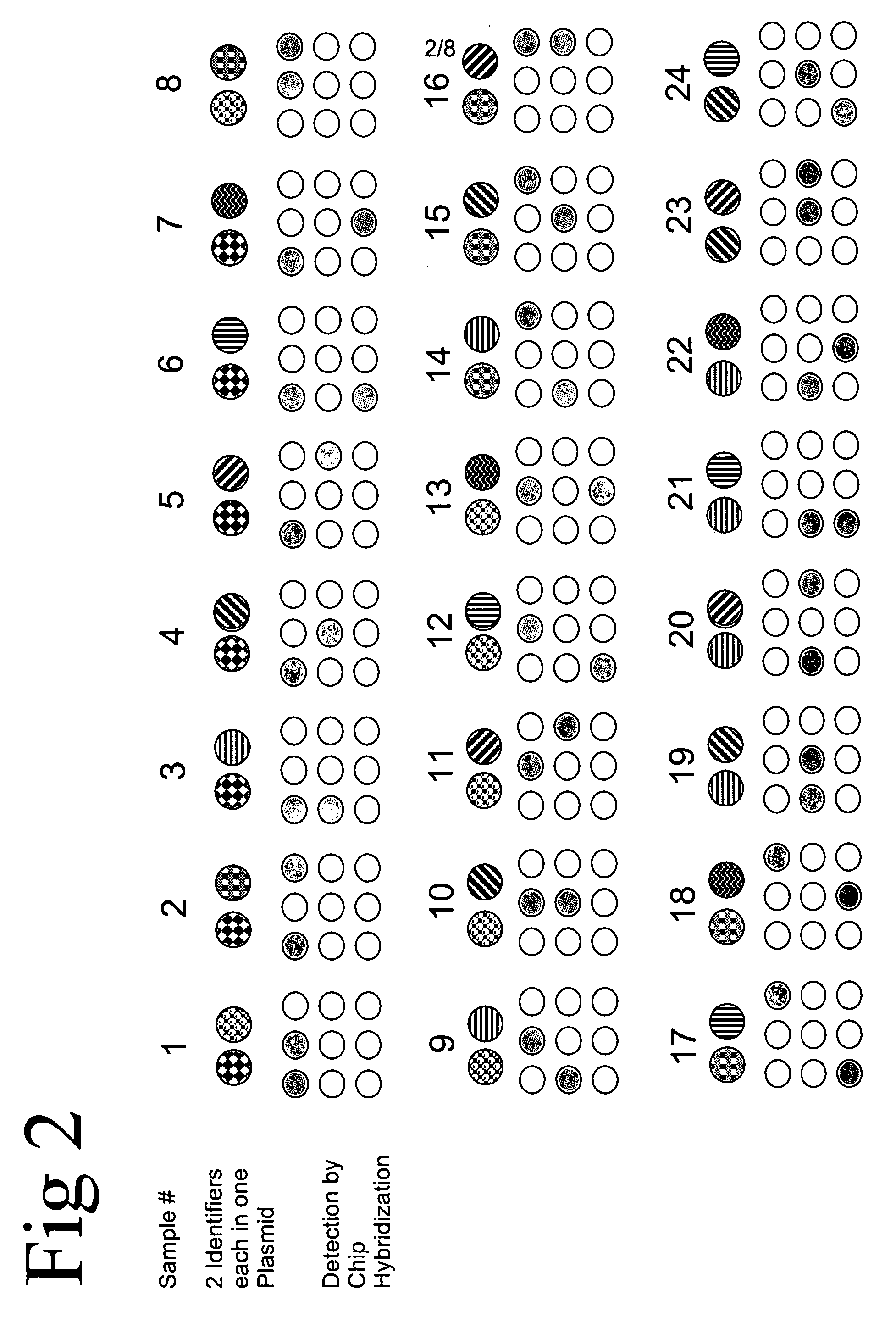
Method of identifying a biological sample for methylation analysis
Aspects of the present invention relate to compositions and methods of identifying at least one biological sample in the field of methylation analysis. In particular aspects at least one biological sample is provided, at least one identifier is applied for each sample, the applied identifier(s) are detected or quantified, and the methylation analysis is performed. Additional aspects provide a methods for testing an experimental procedure. Additional aspects provide kits suitable for realizing the aspects of the invention.
Owner:EPIGENOMICS AG
High-throughput methods for detecting DNA methylation
InactiveUS20030129602A1Bioreactor/fermenter combinationsBiological substance pretreatmentsDNA methylationHybridization probe
The present invention provides a method of hybridization, differential methylation hybridization (DMH) for high throughput methylation analysis of multiple CpG island loci. DMH utilizes nucleic acid probes prepared from a cell sample to screen numerous CpG dinucleotide rich fragments affixed on a screening array. Positive hybridization signals indicate the presence of methylated sites. Methods of preparing the hybridization probes and screening array are also provided.
Owner:UNIVERSITY OF MISSOURI
Branched alpha-glucan, alpha-glucosyltransferase which forms the glucan, their preparation and uses
ActiveUS20100120710A1High yieldA large amountOrganic active ingredientsCosmetic preparationsD-GlucoseGlucose polymers
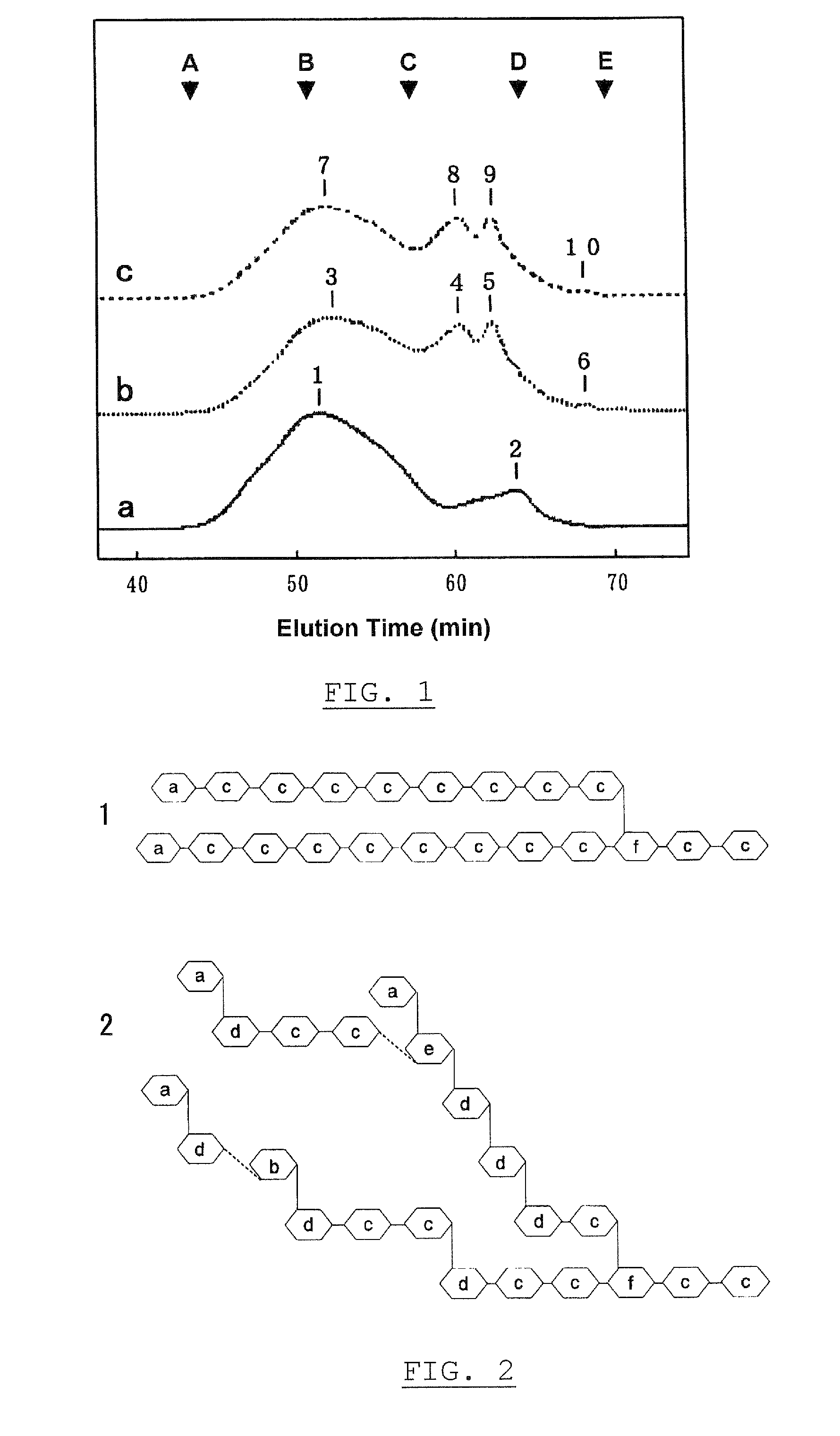
The present invention has objects to provide a glucan useful as water-soluble dietary fiber, its preparation and uses. The present invention solves the above objects by providing a branched α-glucan, which is constructed by glucose molecules and characterized by methylation analysis as follows:(1) Ratio of 2,3,6-trimethyl-1,4,5-triacetyl-glucitol to 2,3,4-trimethyl-1,5,6-triacetyl-glucitol is in the range of 1:0.6 to 1:4;(2) Total content of 2,3,6-trimethyl-1,4,5-triacetyl-glucitol and 2,3,4-trimethyl-1,5,6-triacetyl-glucitol is 60% or higher in the partially methylated glucitol acetates;(3) Content of 2,4,6-trimethyl-1,3,5-triacetyl-glucitol is 0.5% or higher but less than 10% in the partially methylated glucitol acetates; and(4) Content of 2,4-dimethyl-1,3,5,6-tetraacetyl-glucitol is 0.5% or higher in the partially methylated glucitol acetates; a novel α-glucosyltransferase which forms the branched α-glucan, processes for producing them, and their uses.
Owner:HAYASHIBARA CO LTD
Method for methylation analysis
ActiveUS20110009277A1Sugar derivativesMicrobiological testing/measurementMethylation analysisBiology
Aspects of the invention relate to composition and methods for the providing of DNA for methylation analysis that is in particular suitable to be applied in reference laboratories. Further 5 aspects of the invention relate to composition and methods for the highly specific and sensitive methylation analysis of the Septin 9 gene also in particular suitable to be applied in reference laboratories.
Owner:EPIGENOMICS AG
Methods of Analysis of Methylation
InactiveUS20080108073A1Uniformly amplifyStrong specificityMicrobiological testing/measurementMultiplexCytosine
Methods for determining the methylation status of a plurality of cytosines are disclosed. In some aspects genomic DNA target sequences containing CpGs are targeted for analysis by multiplex amplification using target specific probes that can be specifically degraded prior to amplification. The targets may be modified with bisulfite prior to amplification. In another aspect targets are cut with methylation sensitive or insensitive restriction enzymes and marked with a tag using the target specific probes. The presence or absence of methylation may be determined using methylation sensitive restriction enzyme or bisulfite treatment. Detection in many embodiments employs hybridization to tag arrays, genotyping arrays or resequencing arrays.
Owner:AFFYMETRIX INC
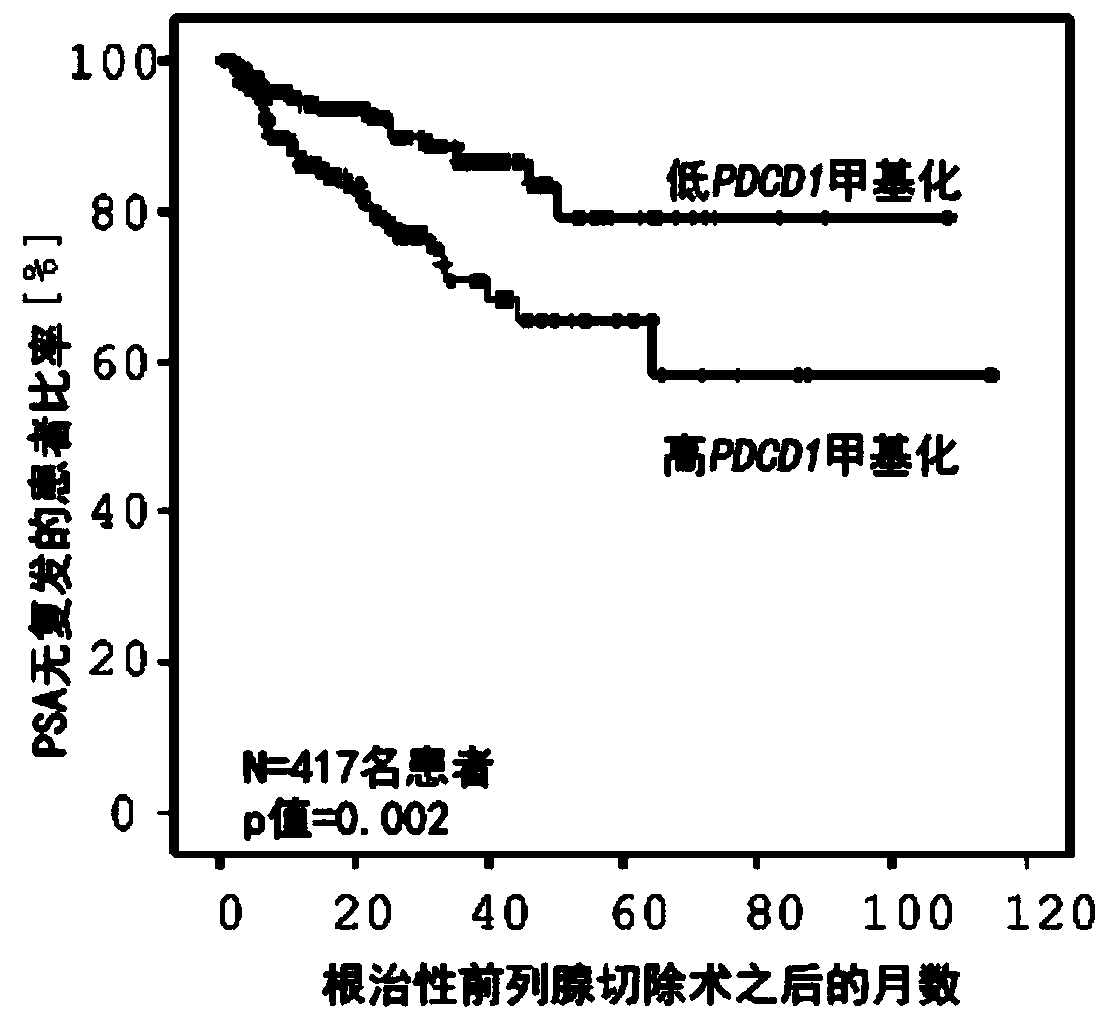
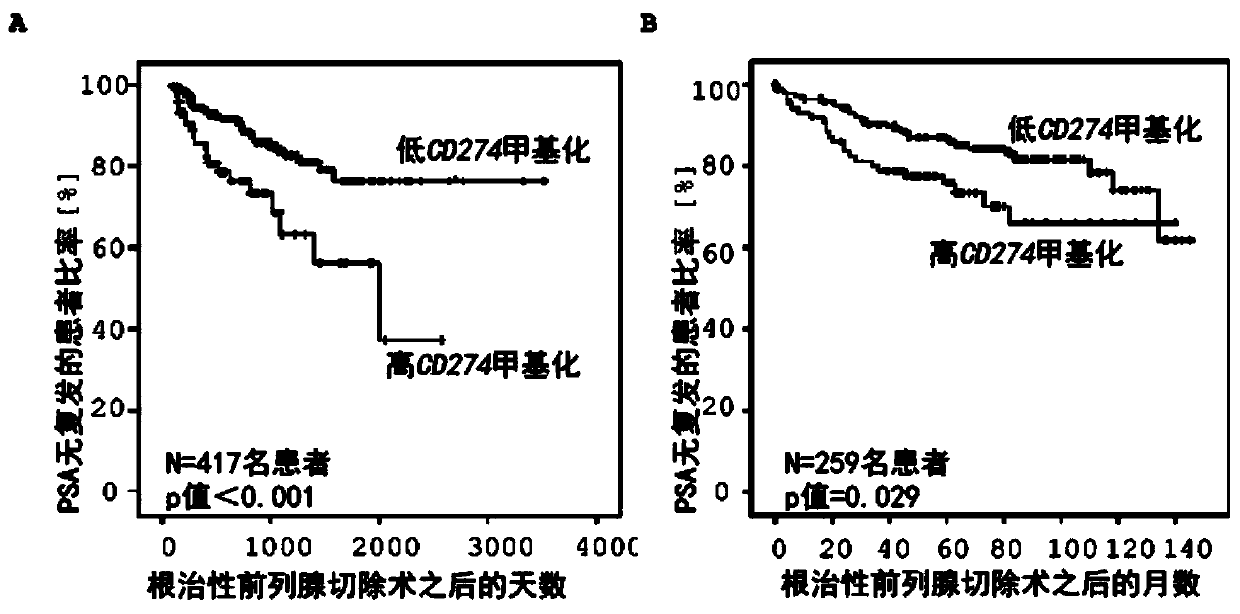
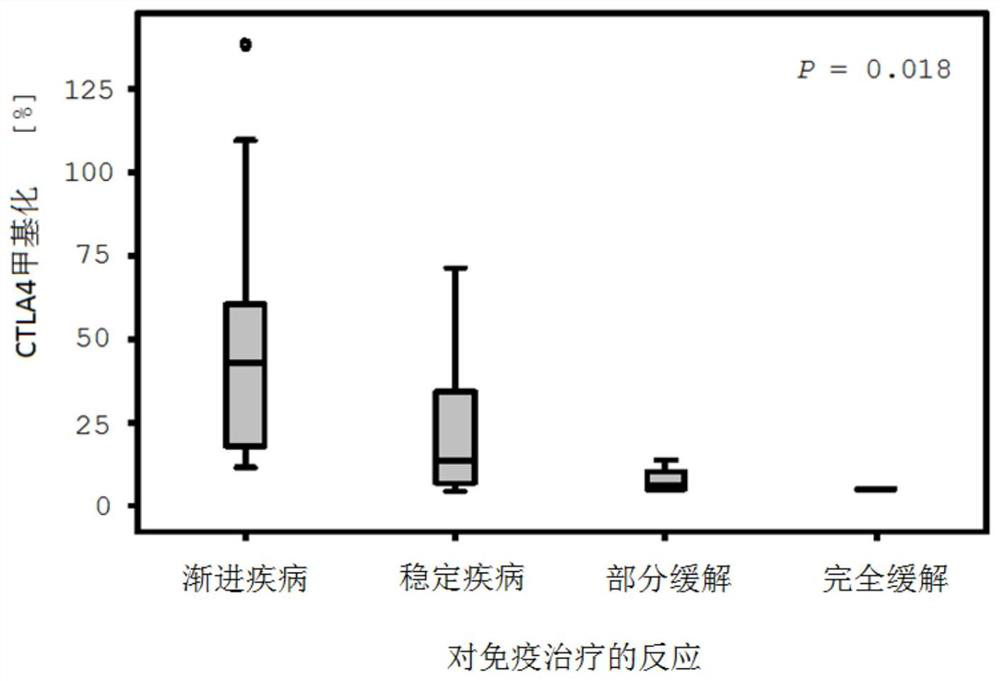
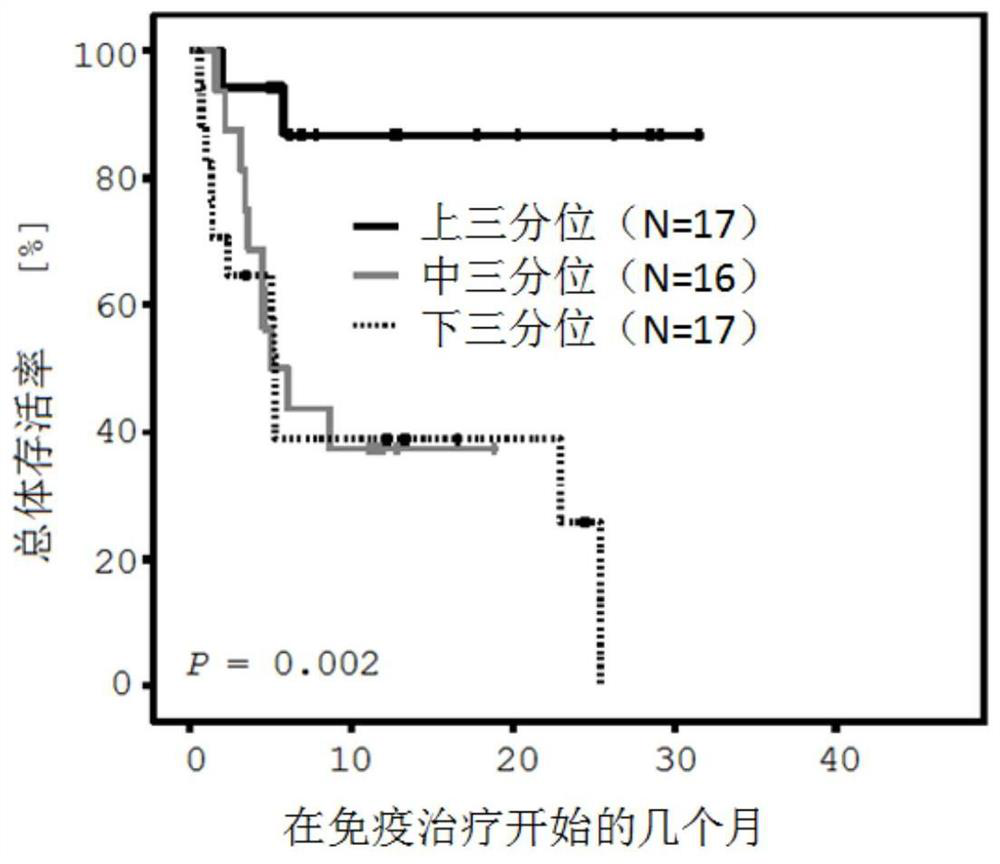
Method for assessing a prognosis and predicting the response of patients with malignant diseases to immunotherapy
The invention relates to, among others, a method for assessing a prognosis of a patient with a malignant disease and / or for predicting the response of a patient with a malignant disease to immunotherapy. For this purpose, a DNA methylation analysis is carried out on at least one immunoregulatory gene of cells of the malignant disease and / or T lymphocytes which interact with the cells of the malignant disease, said gene coding for an immune checkpoint selected from B7 proteins and the receptors thereof, MHC-peptide complex-binding co-receptors, the members of the tumor necrosis factor receptorsuperfamily TNFRSF9, CD40, TNFRSF4, TNFRSF18, and CD27, the members of the immunoglobulin superfamily TIGIT, BTLA, HAVCR2, BTNL2, and CD48, and the andenosine-binding adenosine 2A receptor.
Owner:迪莫迪特里希
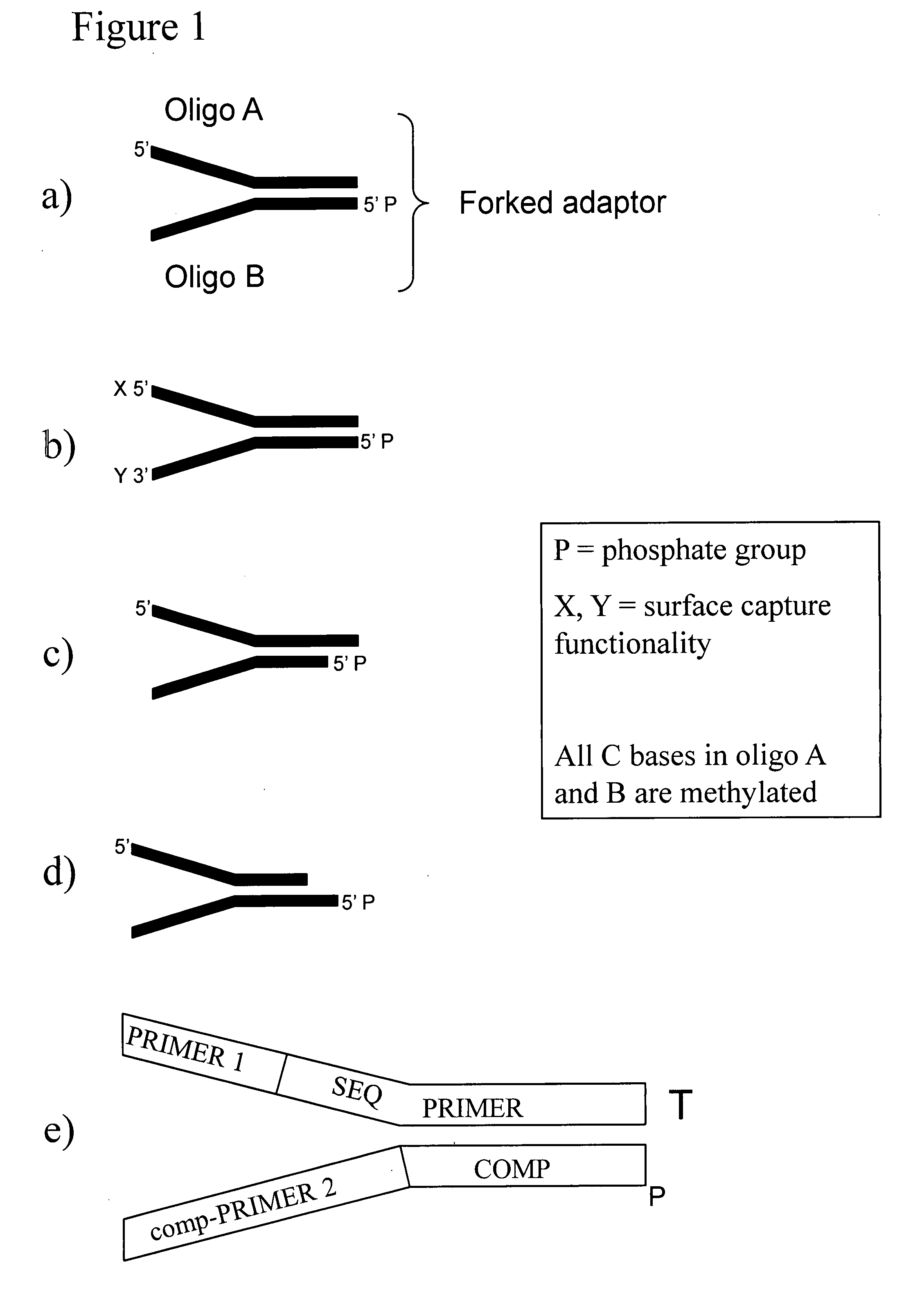
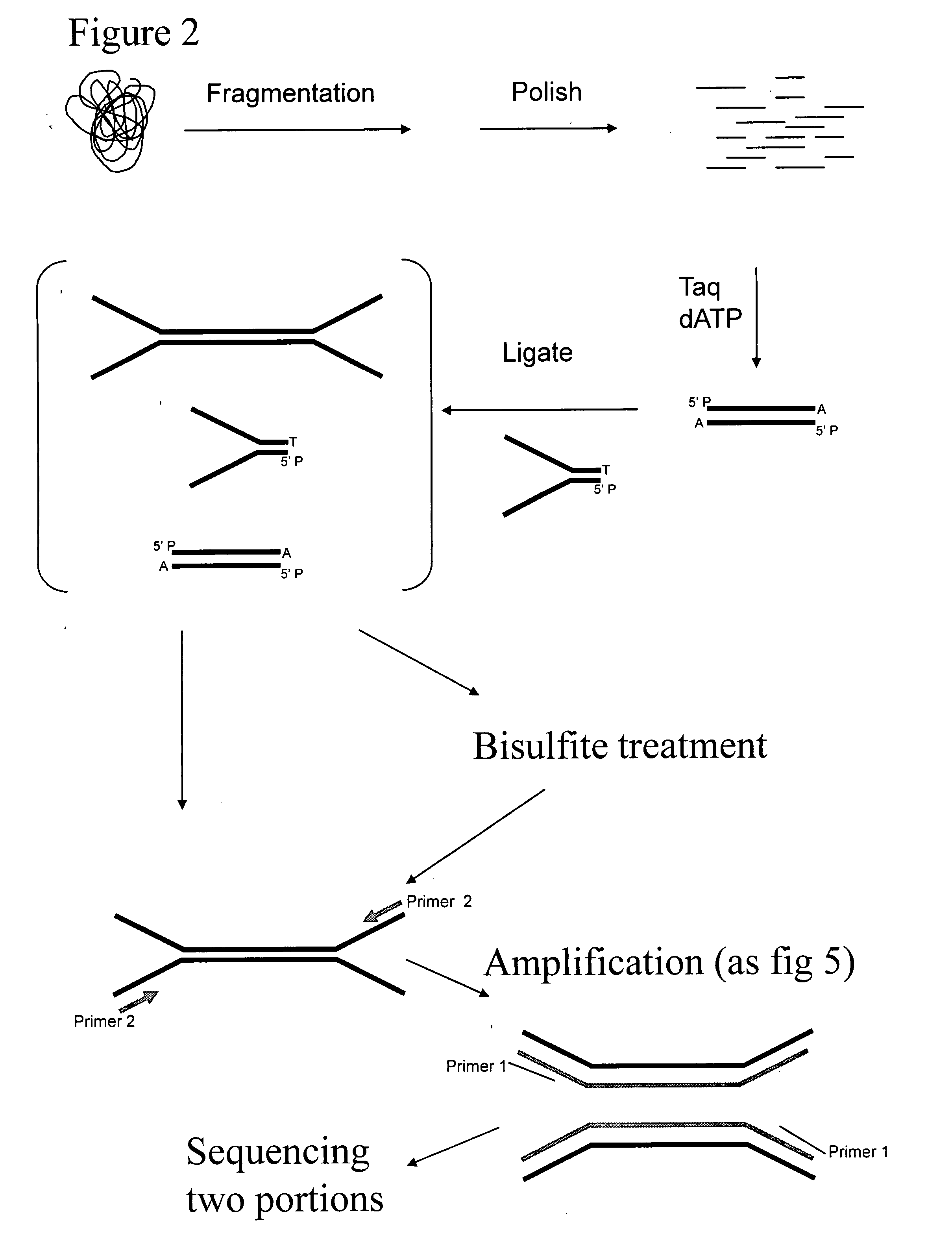
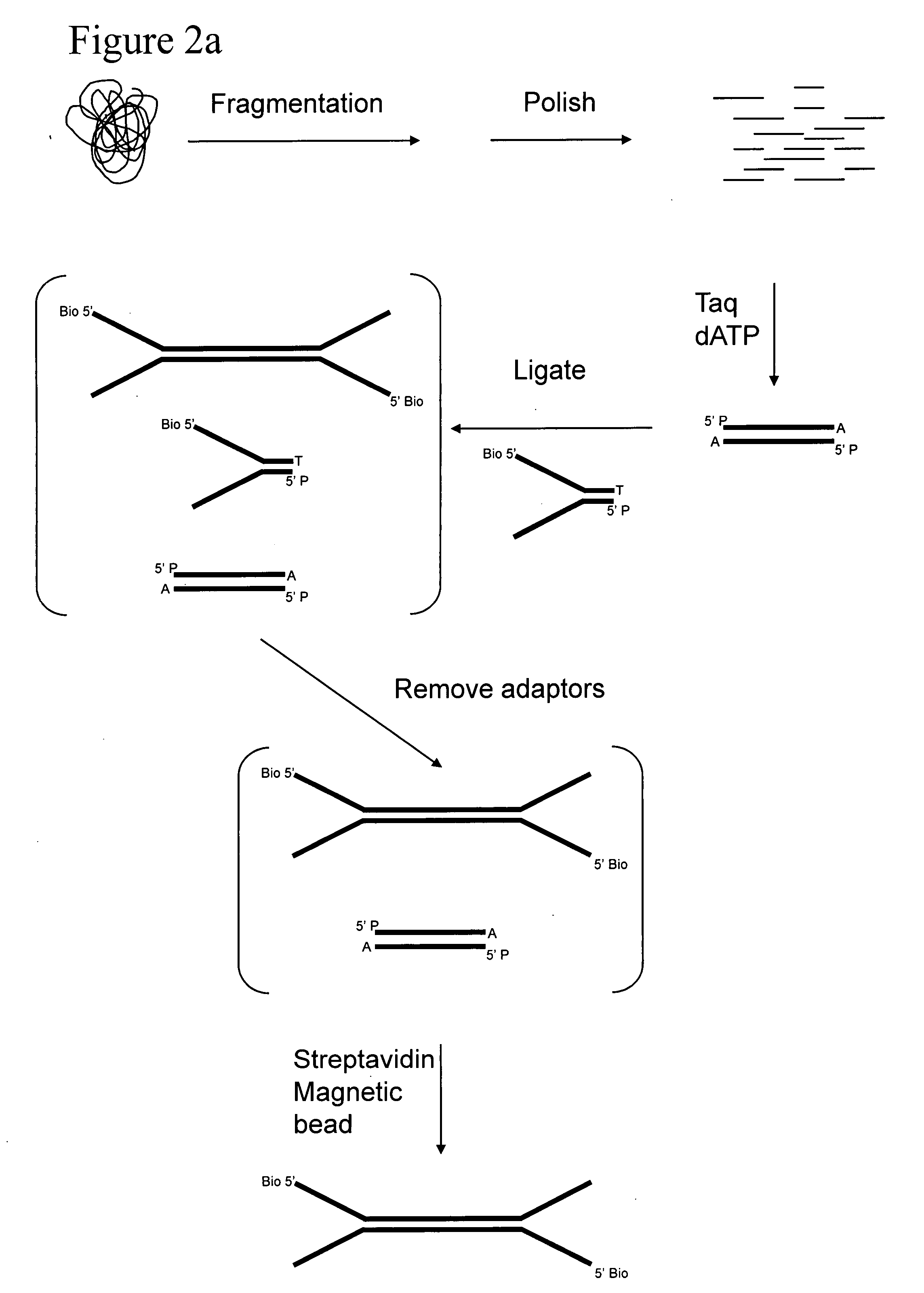
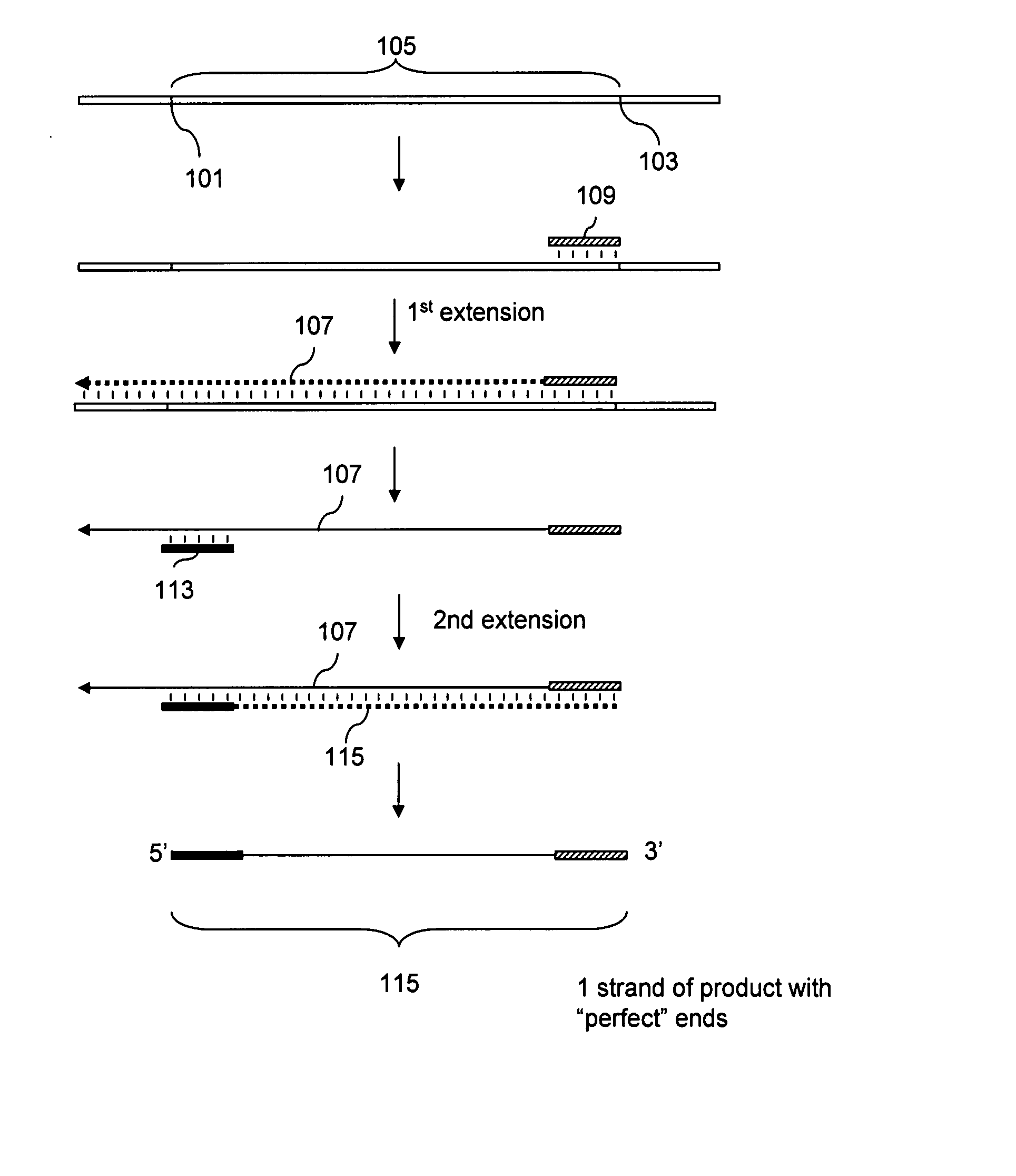
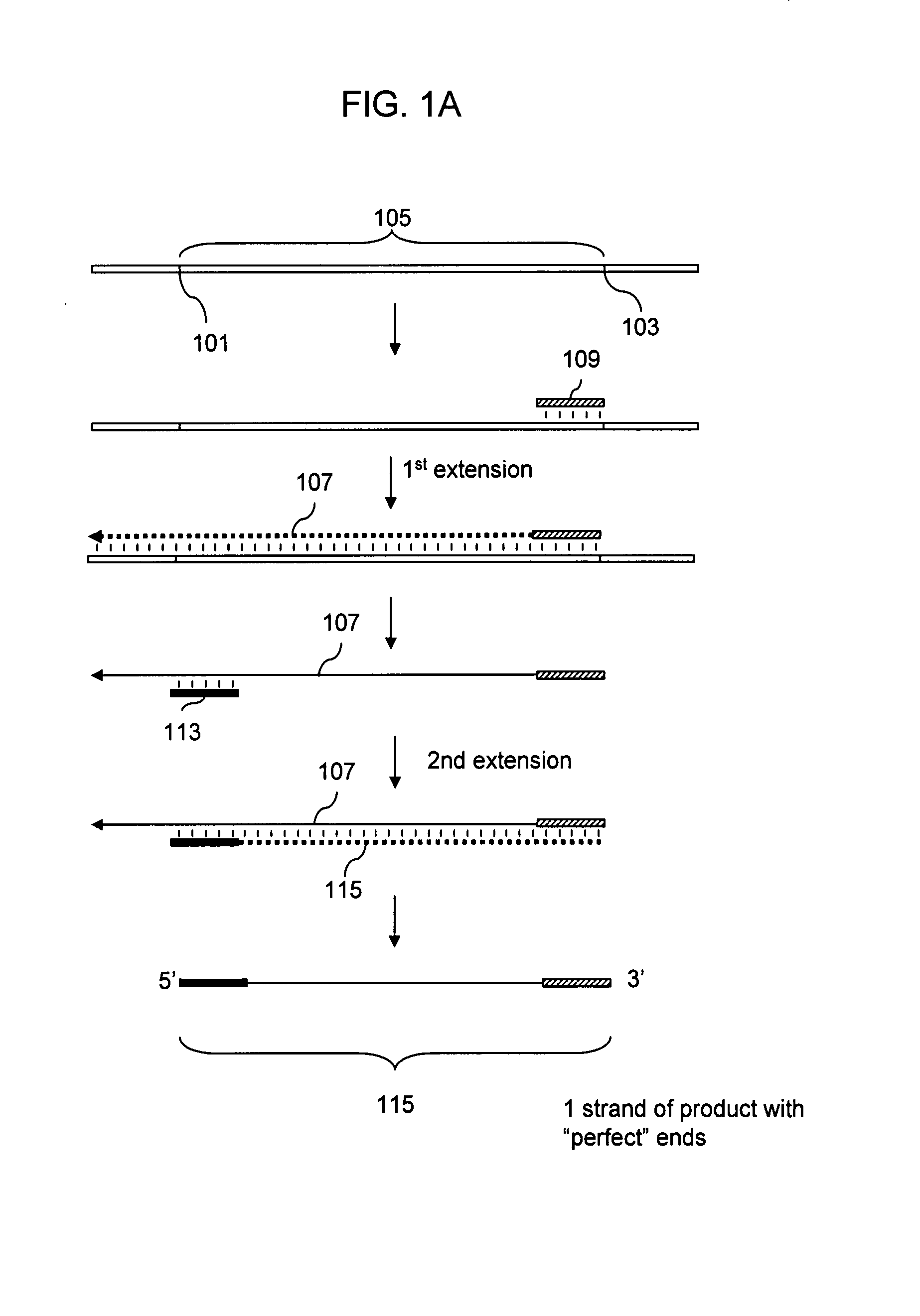
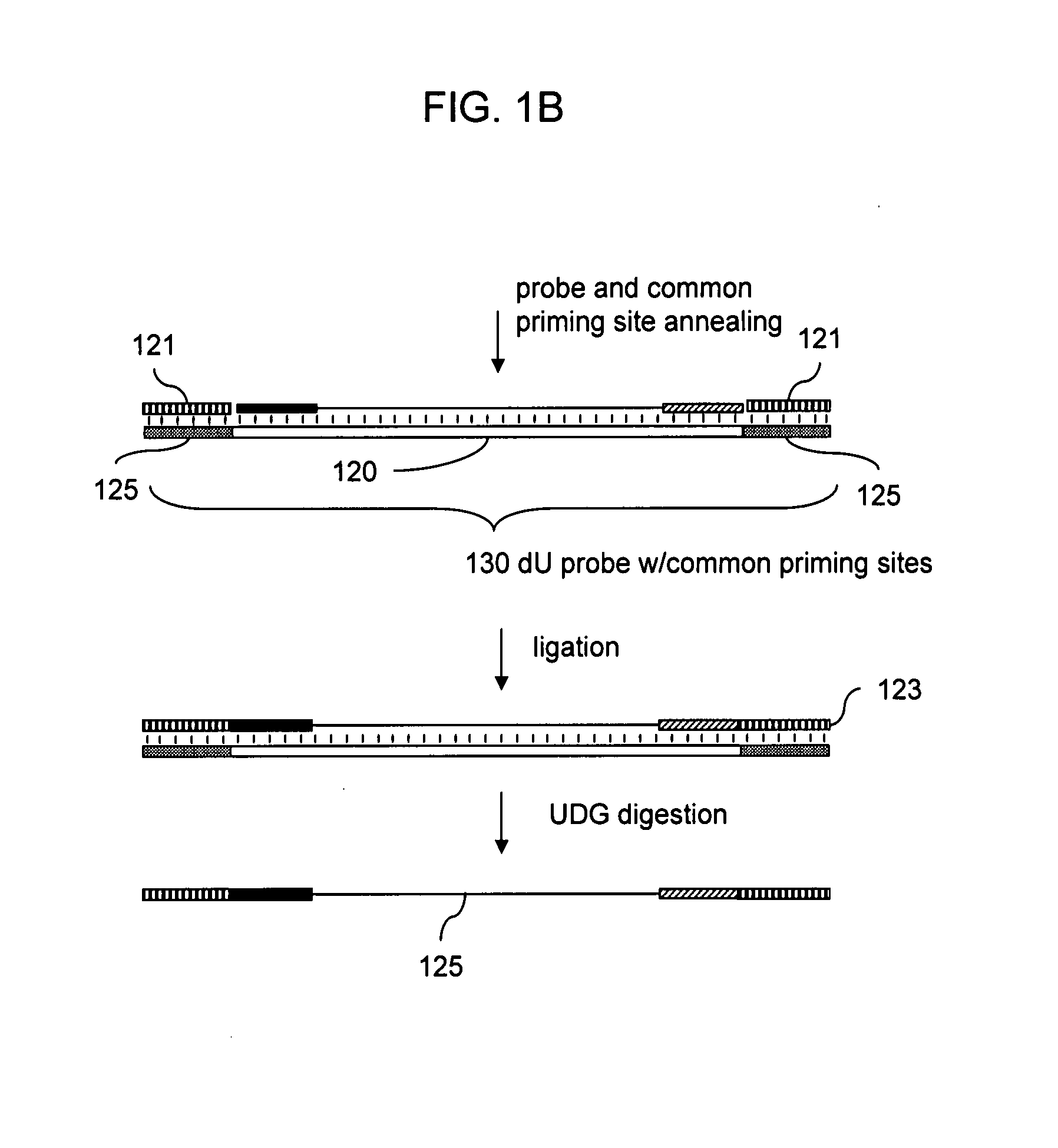
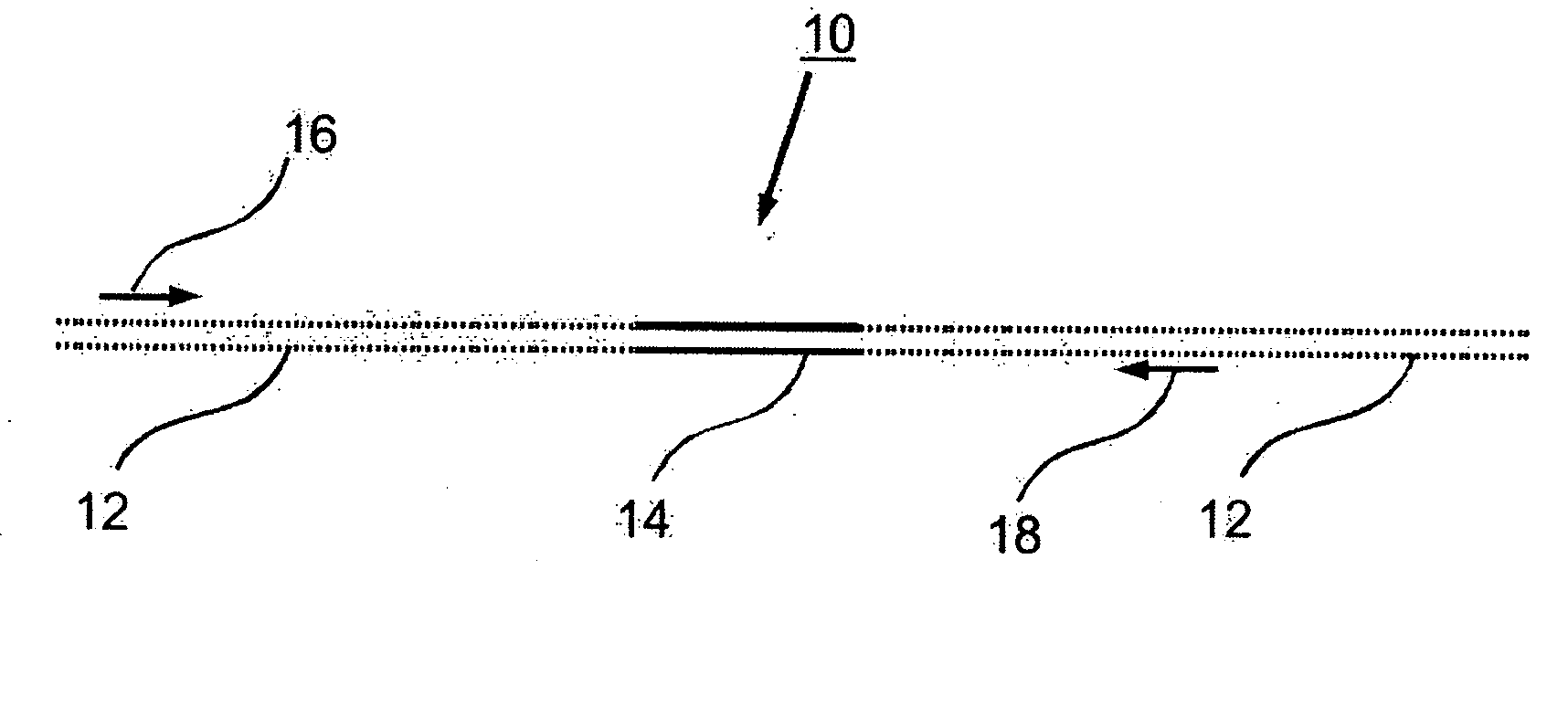
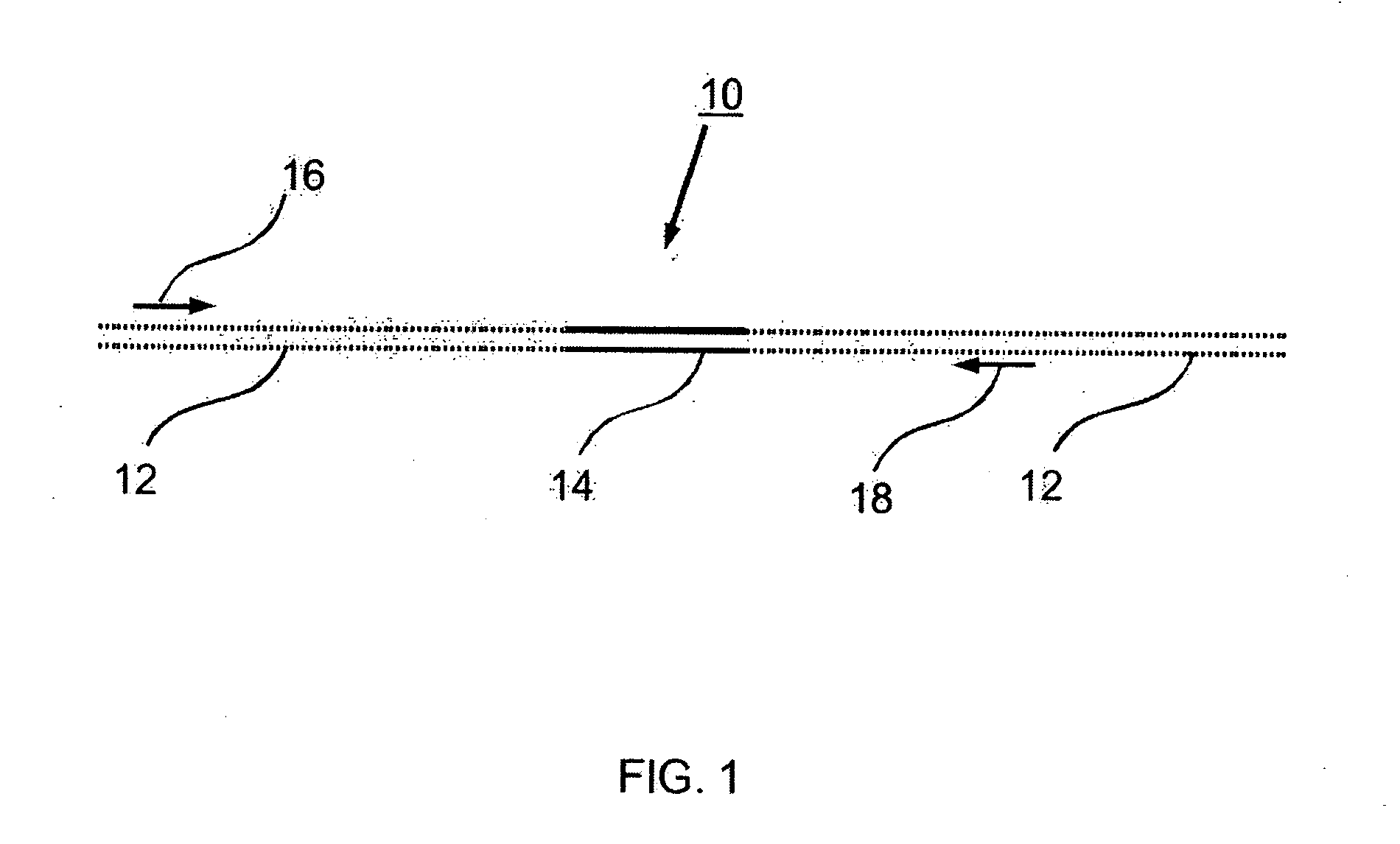
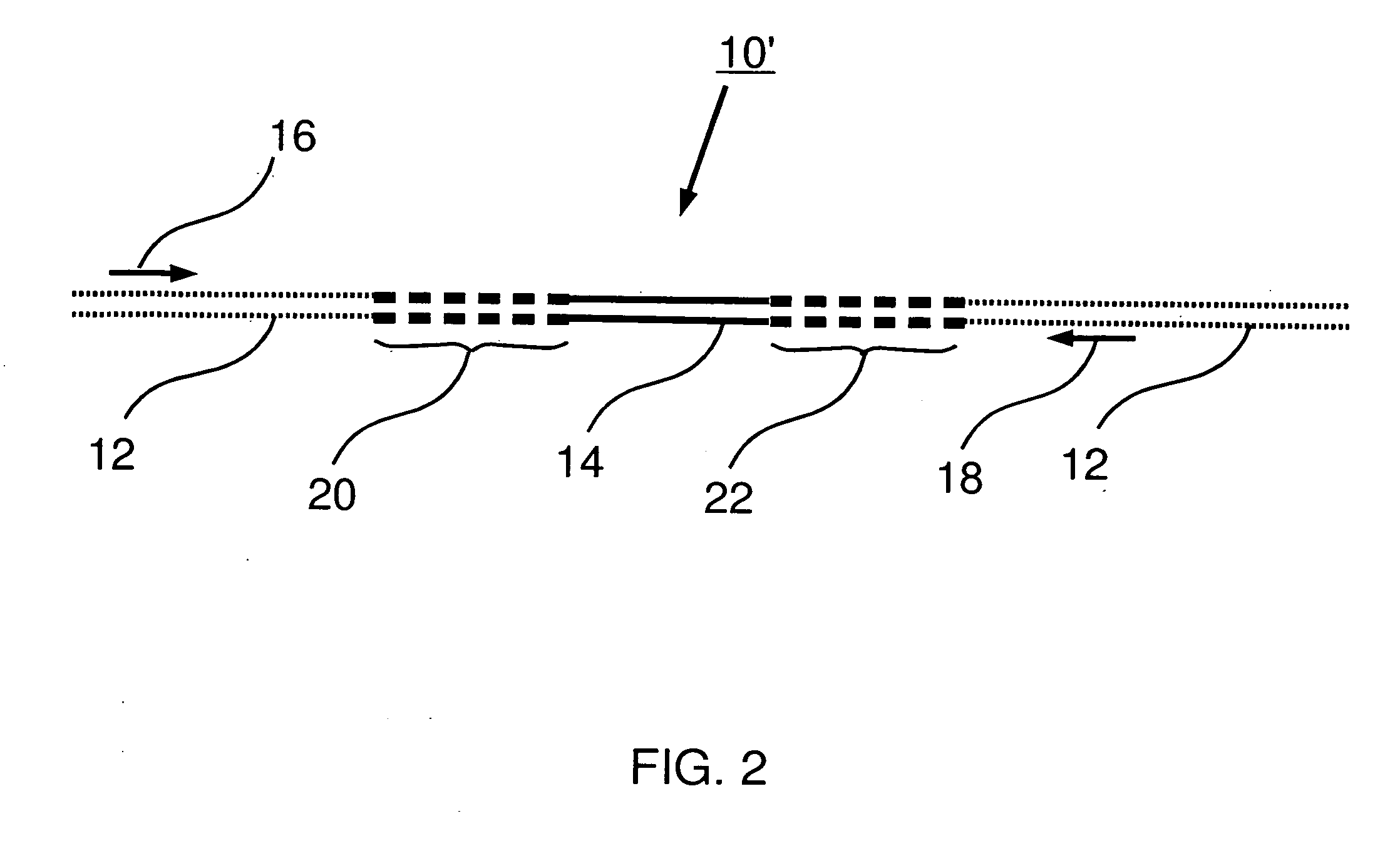
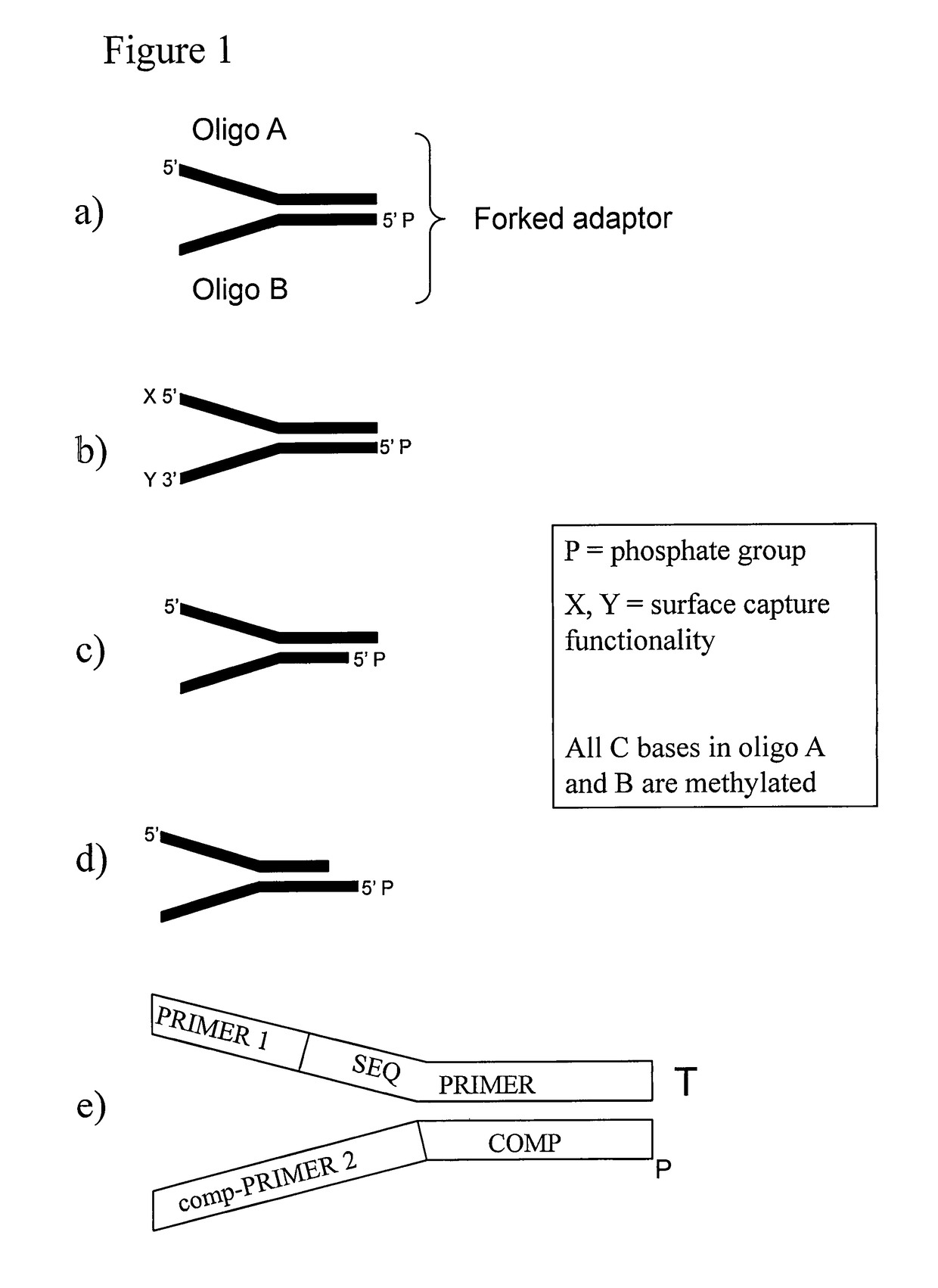
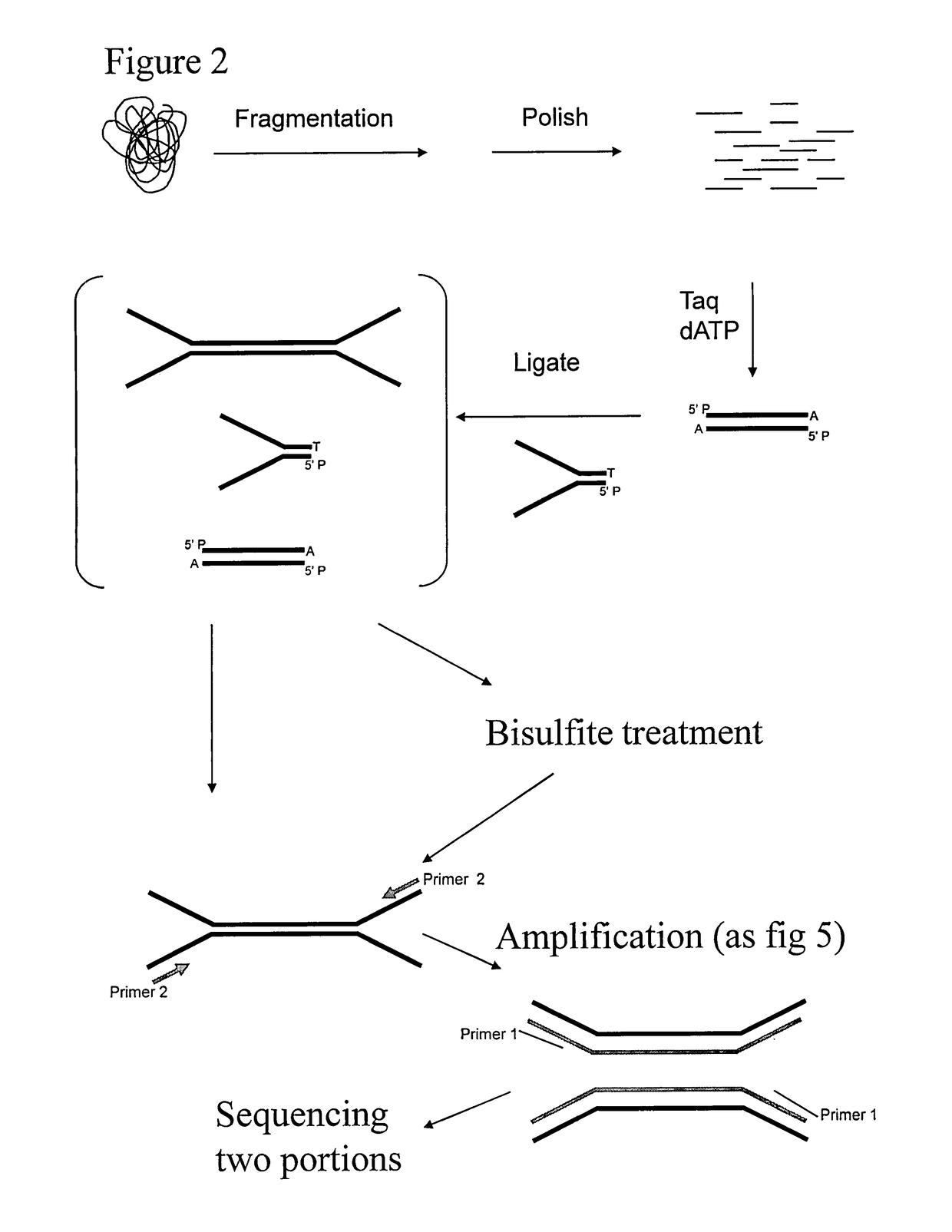
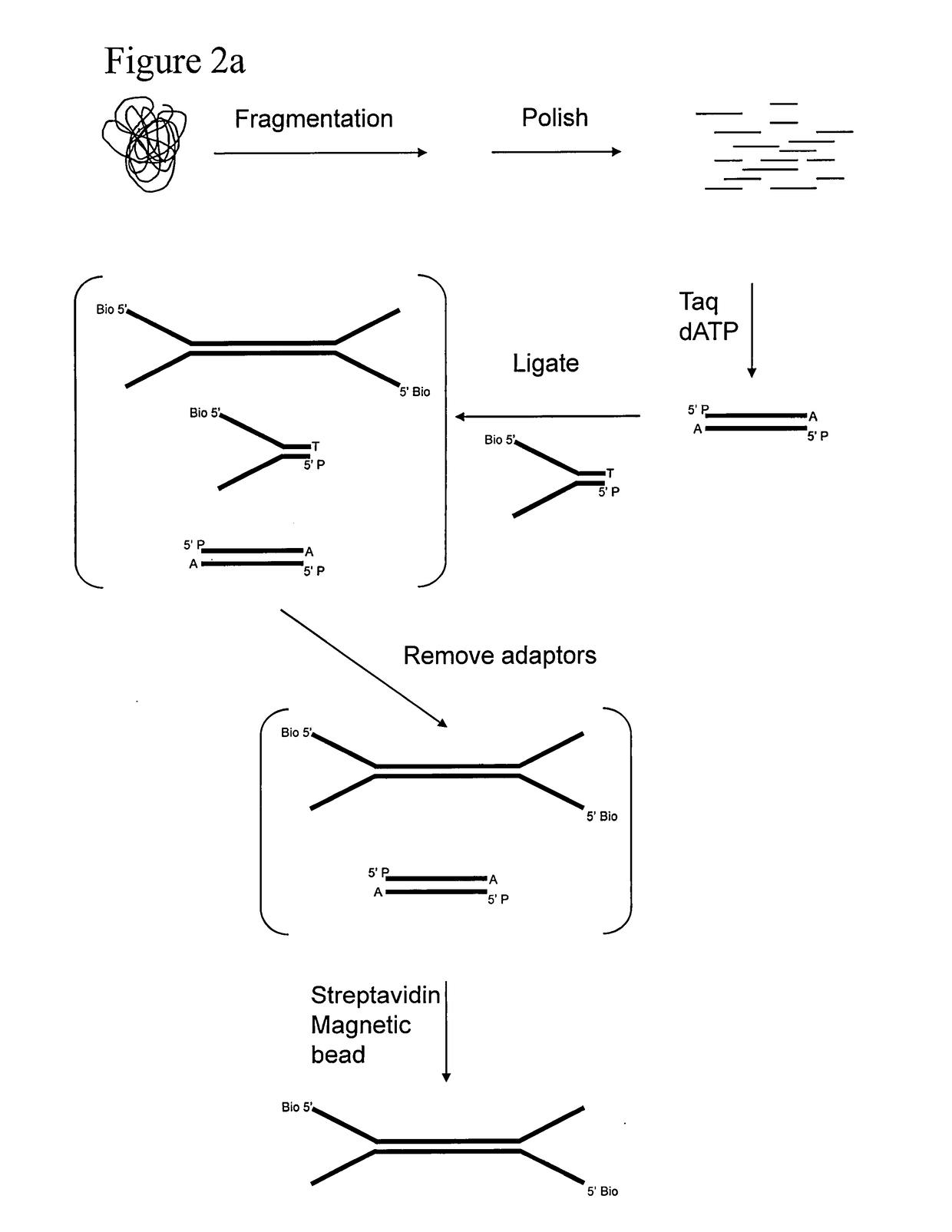
Preparation of templates for methylation analysis
ActiveUS20090148842A1Convenient treatmentEasy to analyzeMicrobiological testing/measurementCytosineNucleotide
The invention relates to a method of preparing and using a library of template polynucleotides suitable for use as templates in solid-phase nucleic acid amplification and sequencing reactions to determine the methylation status of the cytosine bases in the library. In particular, the invention relates to a method of preparing and analysing a library of template polynucleotides suitable for methylation analysis.
Owner:MASSACHUSETTS INST OF TECH +1
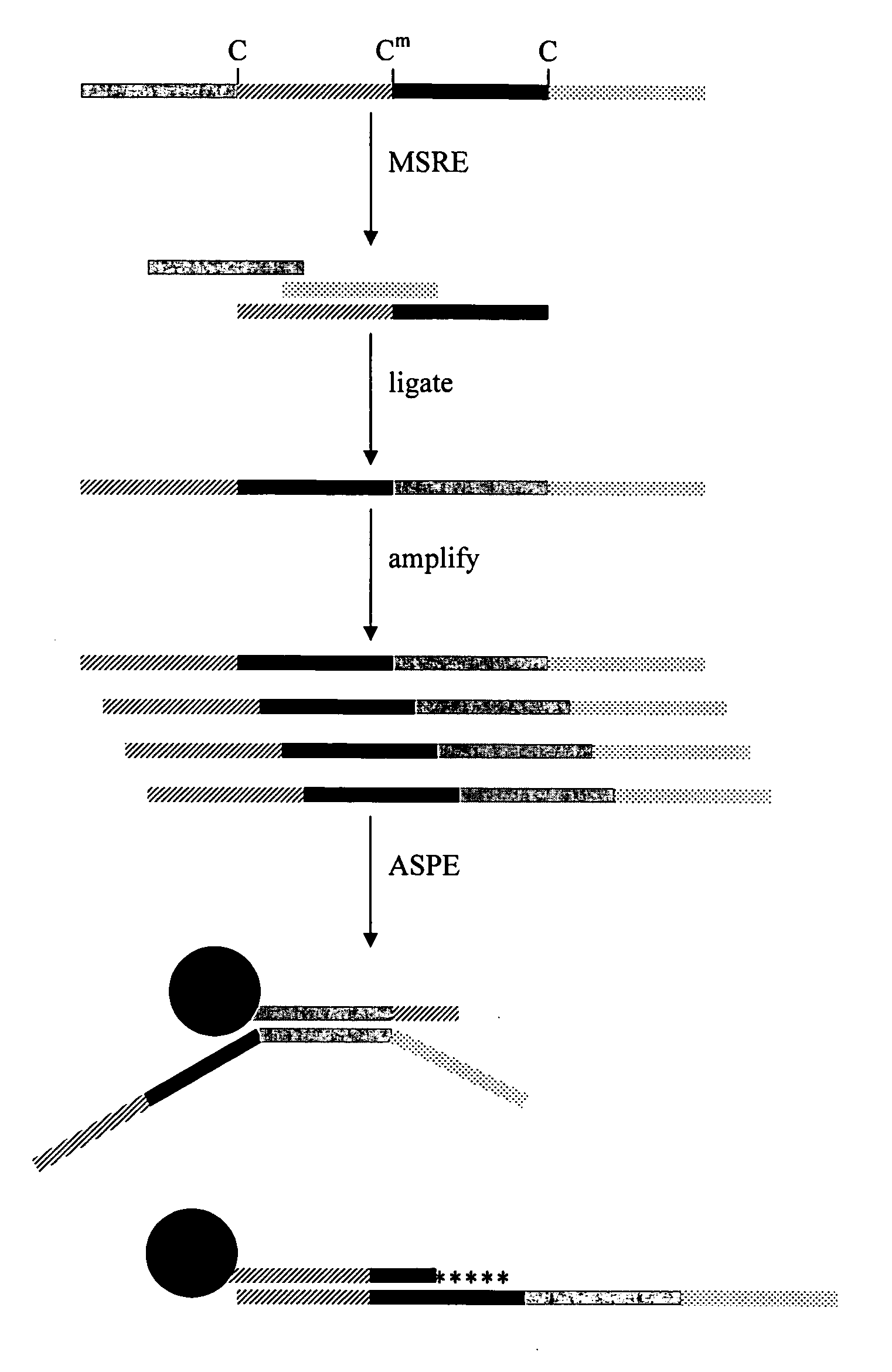
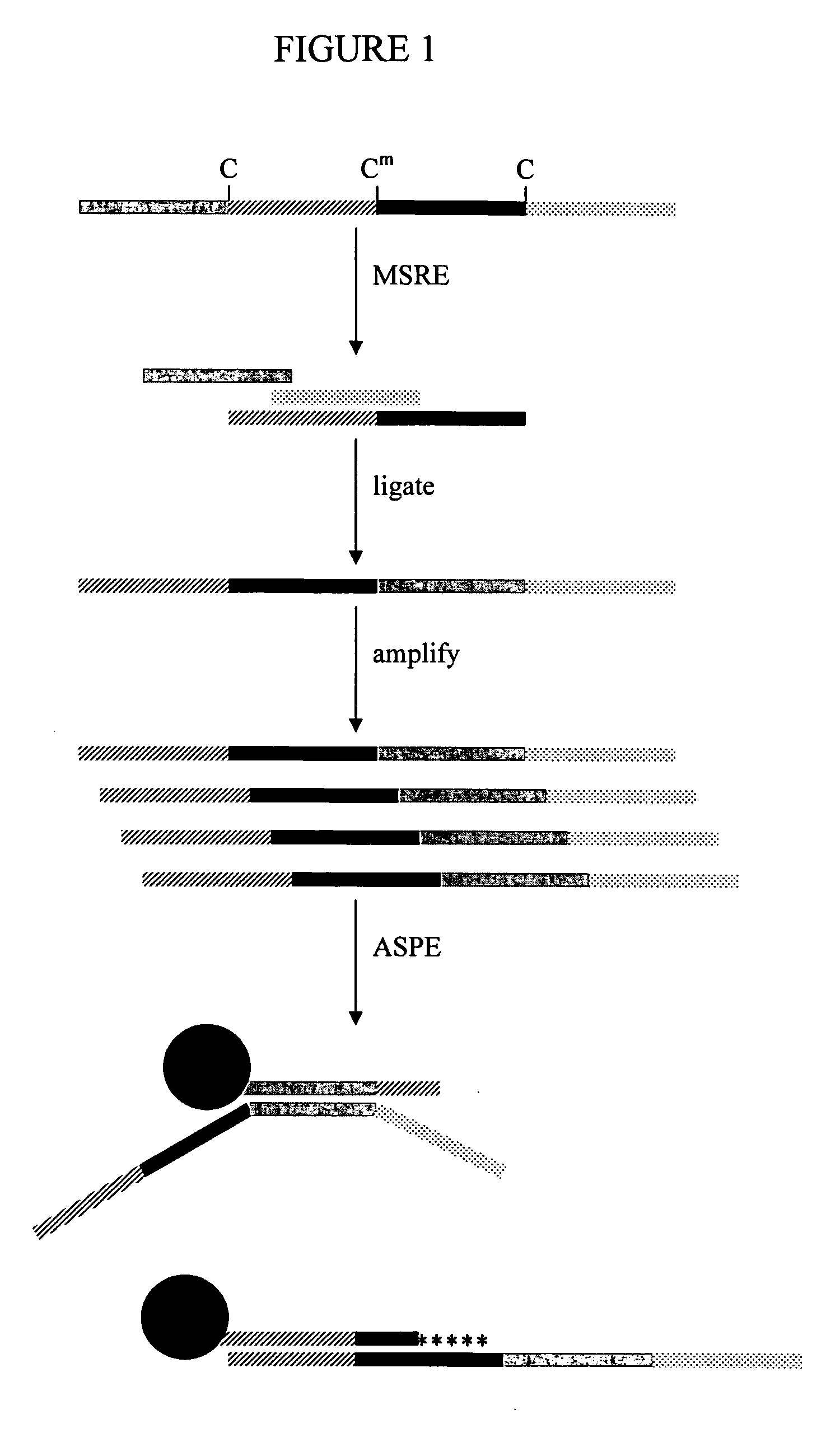
Methylation-sensitive restriction enzyme endonuclease method of whole genome methylation analysis
The invention provides methods of identifying a plurality of reactive recognition sites for a restriction endonuclease in genomic DNA. In particular embodiments, the methods can be used to identify methylation state of a plurality of CpG target sites in genomic DNA. The method can include steps of treating genomic DNA with a restriction endonuclease, thereby producing genomic DNA fragments; ligating the fragments, thereby forming a concatenated DNA; and identifying sequence portions of the concatenated DNA that are re-ordered compared to the genomic DNA.
Owner:ILLUMINA INC
Methods of Analysis of Methylation
InactiveUS20110151438A9Uniformly amplifyStrong specificityMicrobiological testing/measurementMultiplexCytosine
Owner:AFFYMETRIX INC
Methods for preservation of genomic DNA sequence complexity
ActiveUS20110027789A1Easy diagnosisImproved prognosisSugar derivativesMicrobiological testing/measurementCytosineNucleotide
The present invention relates to methods and kits for preserving genomic DNA sequence complexity within chemically and / or enzymatically converted DNA by an enzyme or series of enzymes that adds a methyl group to a cytosine outside of CpG dinucleotide sequences of genomic DNA. Further, the present invention relates to methylation analysis of the genomic DNA.
Owner:EPIGENOMICS AG
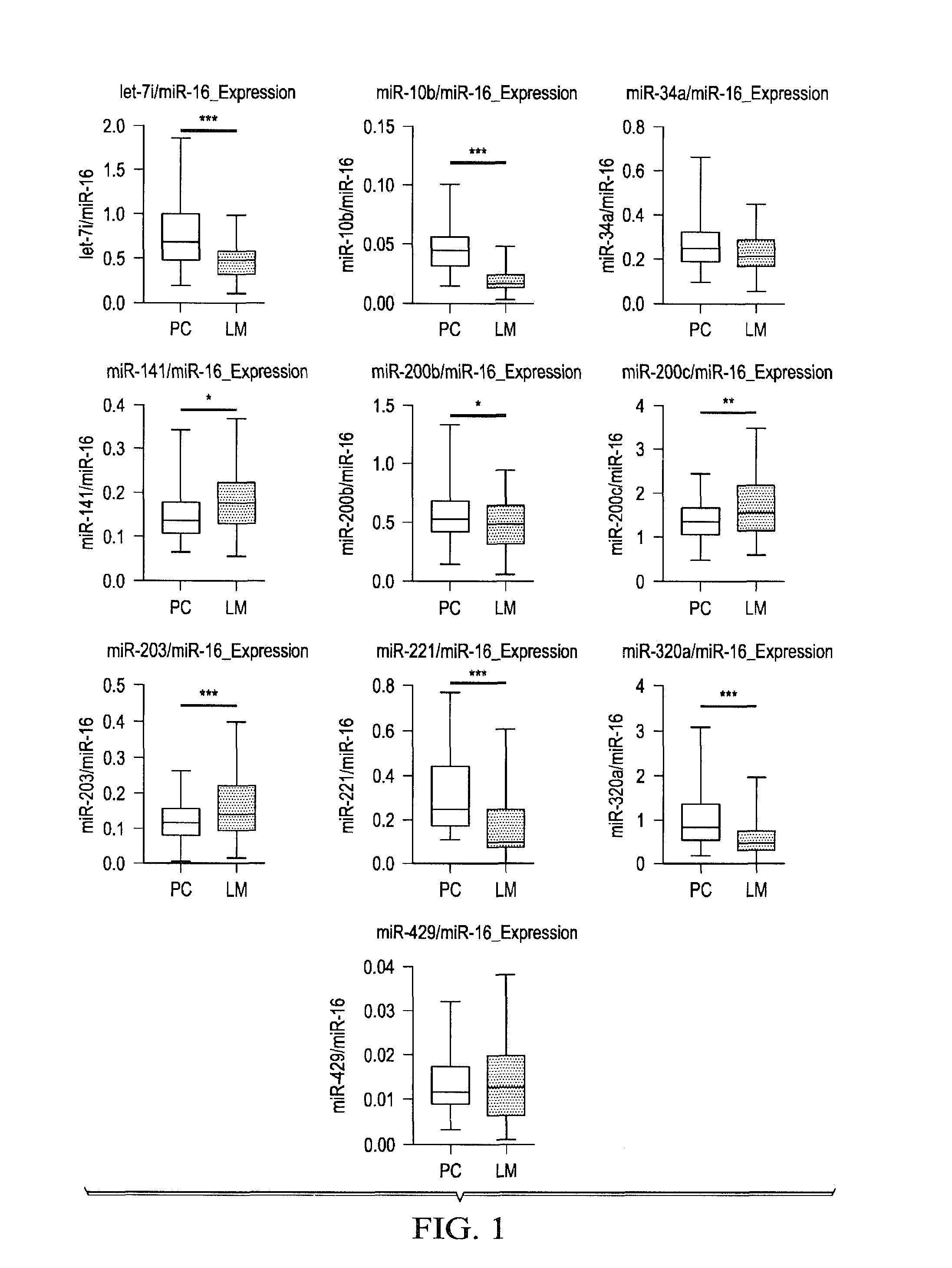
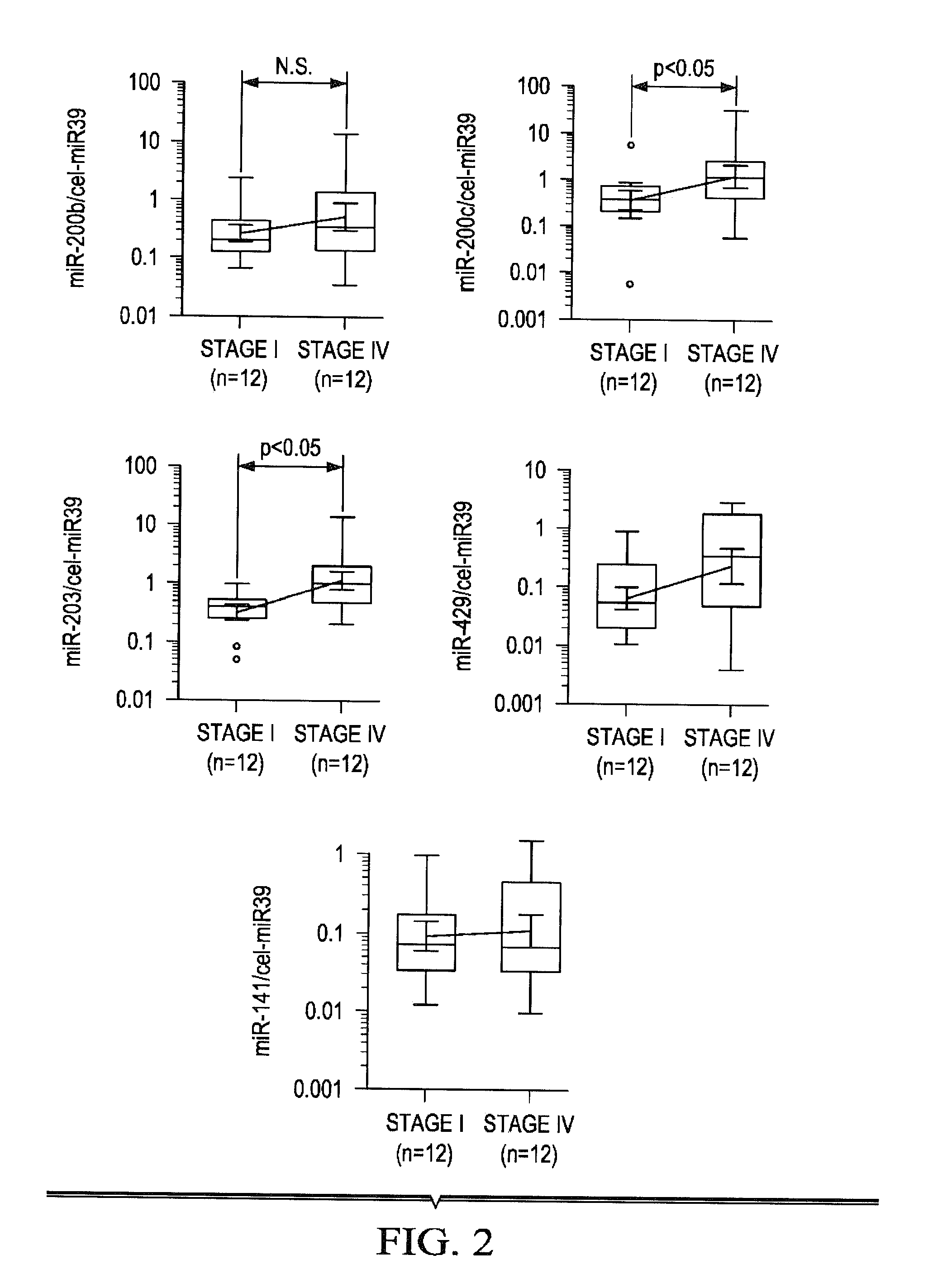
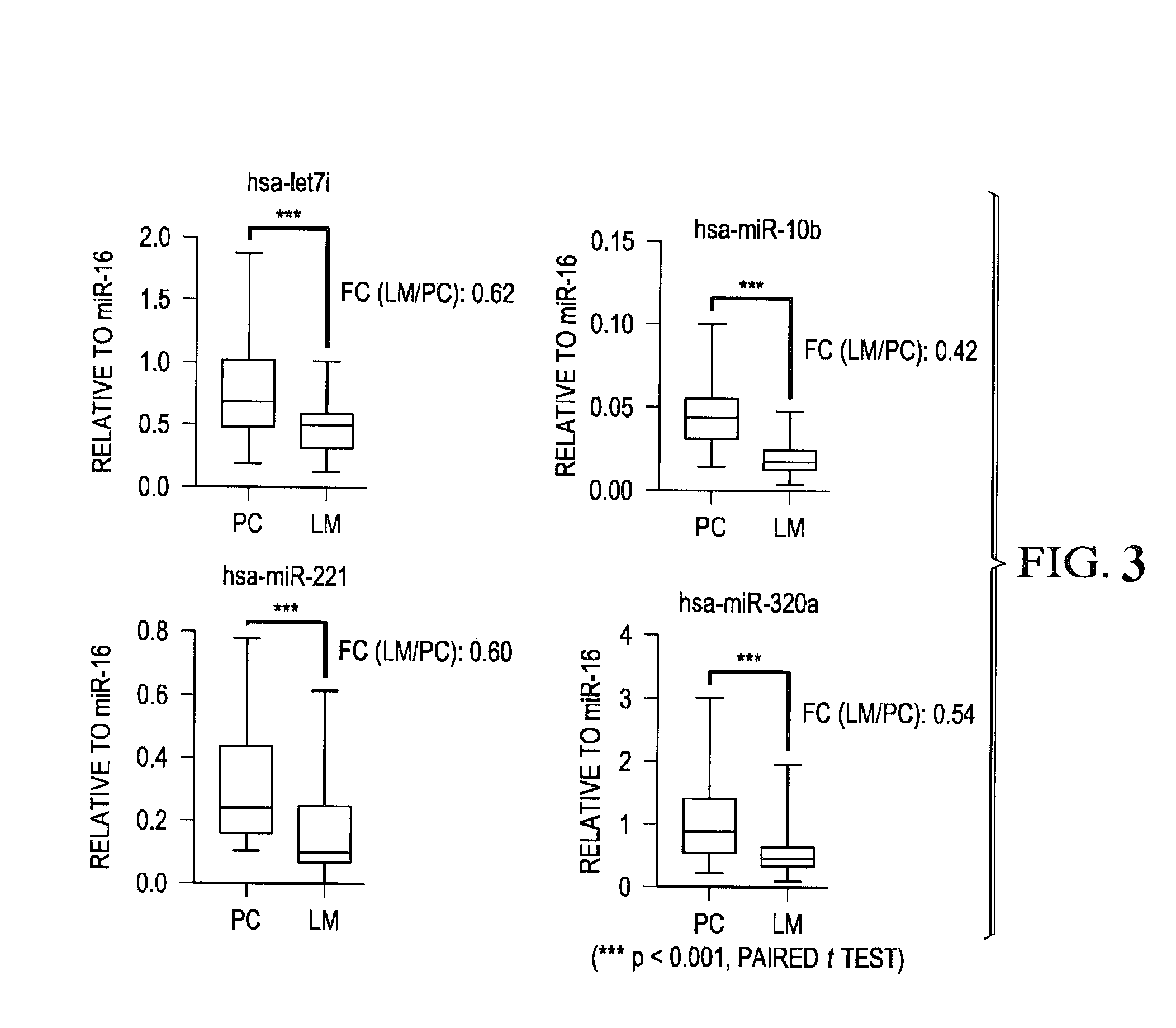
Tissue & blood-based mirna biomarkers for the diagnosis, prognosis and metastasis-predictive potential in colorectal cancer
ActiveUS20140322354A1Heavy metal active ingredientsMicrobiological testing/measurementOncologyMethylation analysis
Methods and compositions for the diagnosis, prognosis and classification of cancer, especially colorectal cancer, are provided. For example, in certain aspects methods for cancer prognosis using expression or methylation analysis of selected biomarkers are described. Particular aspects of the present invention may include methods and biomarkers for diagnosing or detecting colorectal cancer or metastasis in a subject by measuring a level of expression of biomarker miRNA such as miR-885-5p in the sample from the subject and evaluating the risk of developing cancer or metastasis in the subject.
Owner:BAYLOR RES INST
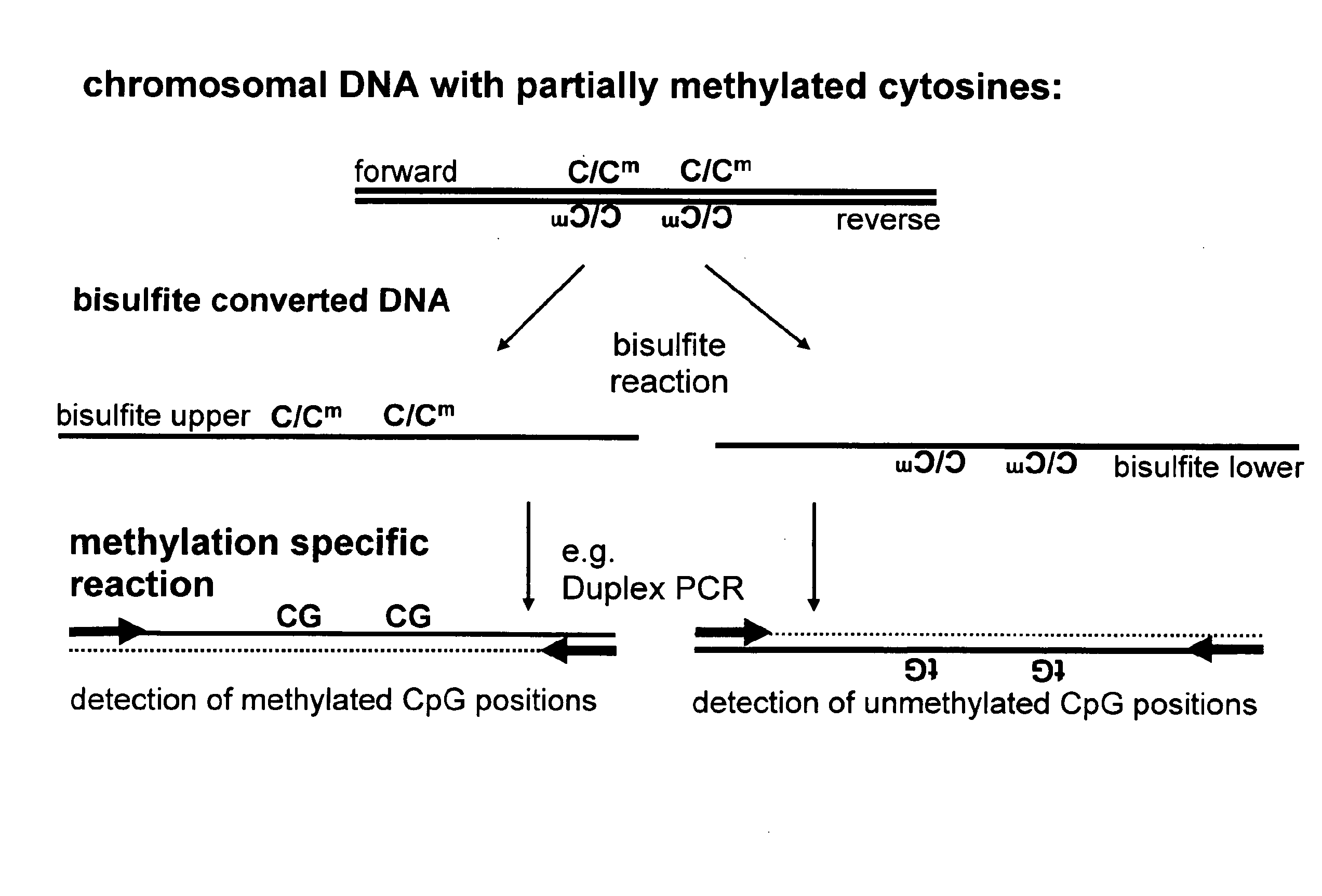
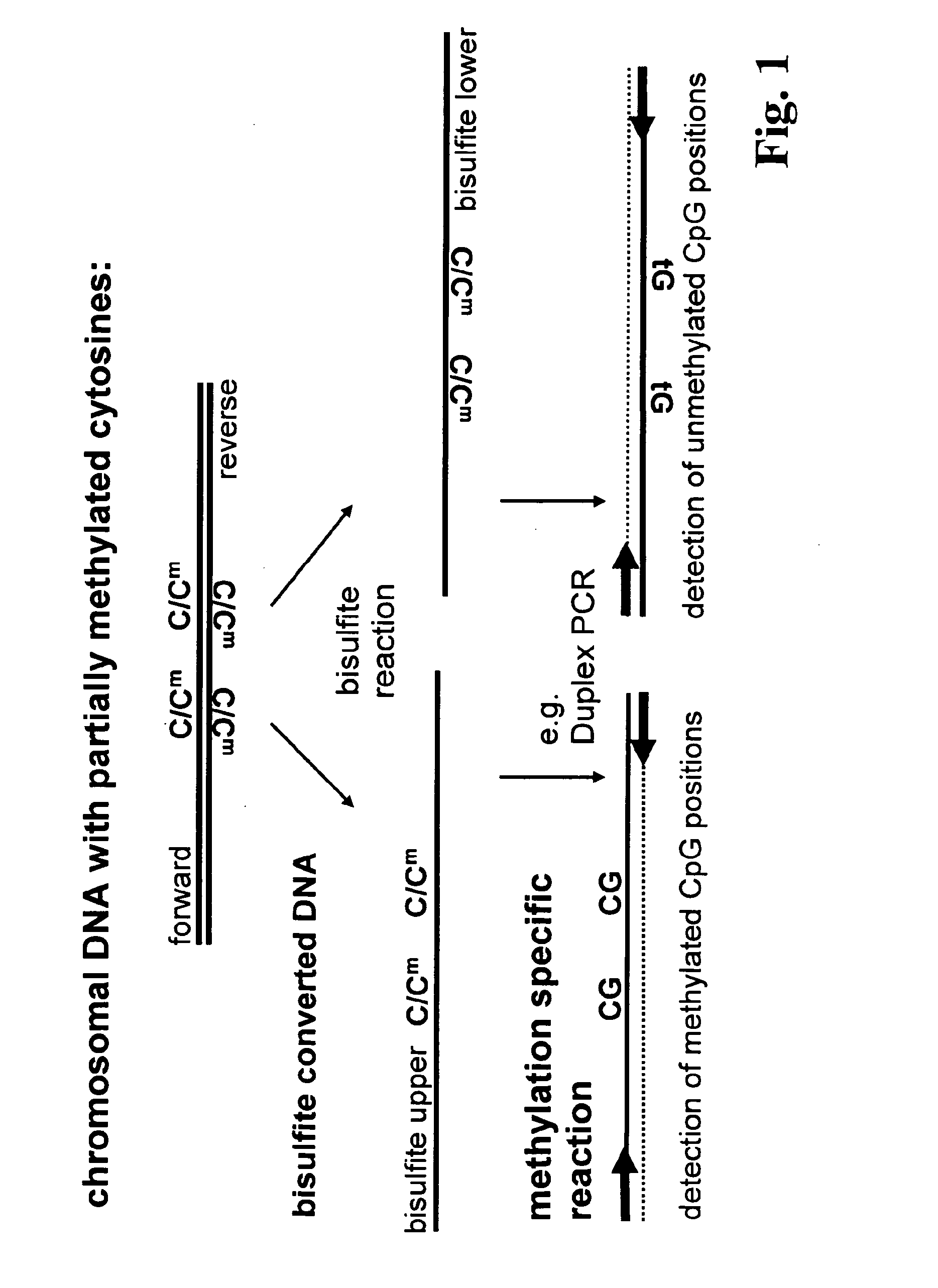
Method for methylation analysis of nucleic acid
ActiveUS20100092951A1High sensitivityMicrobiological testing/measurementBiological testingBiochemistryMethylation analysis
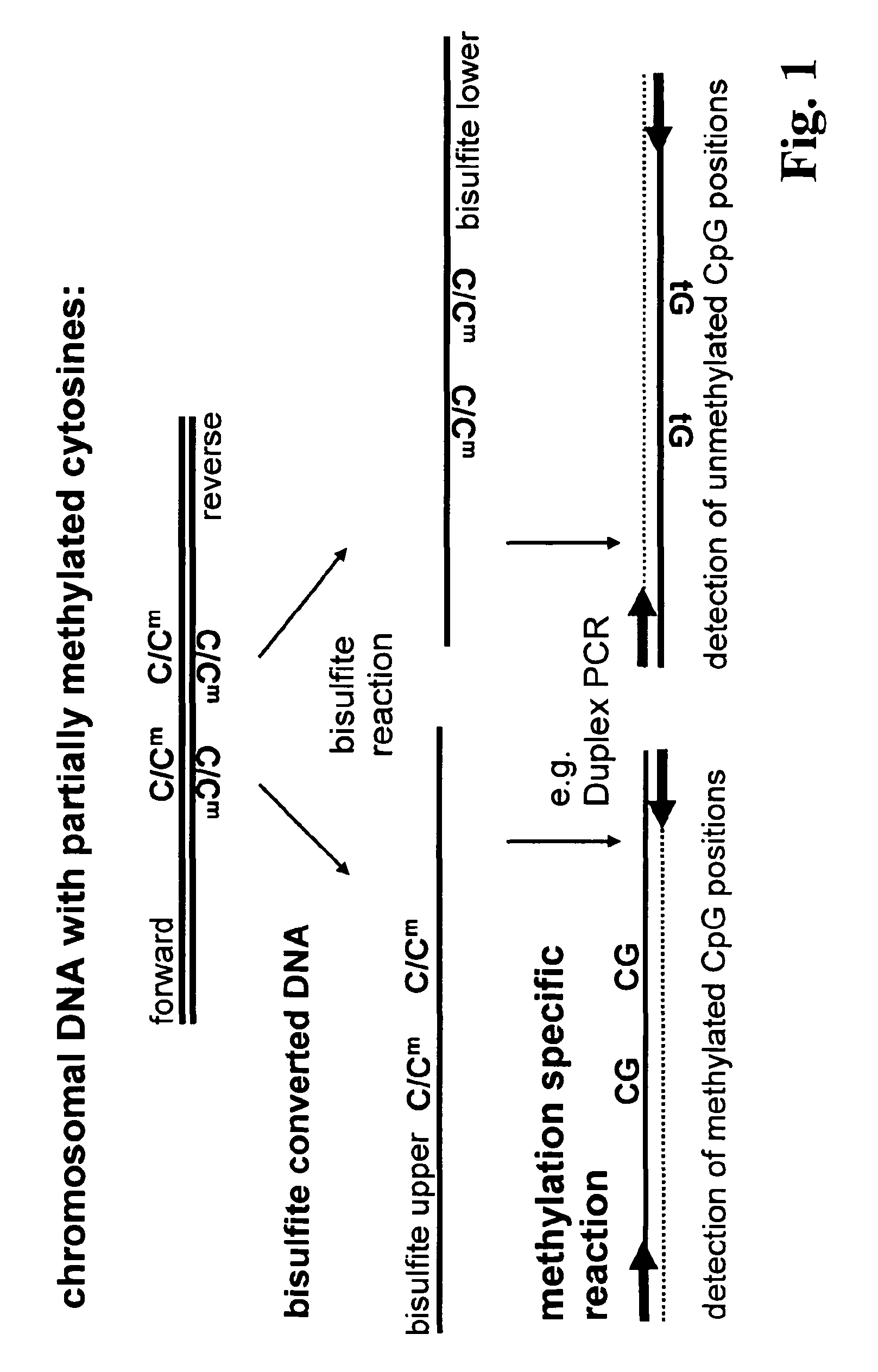
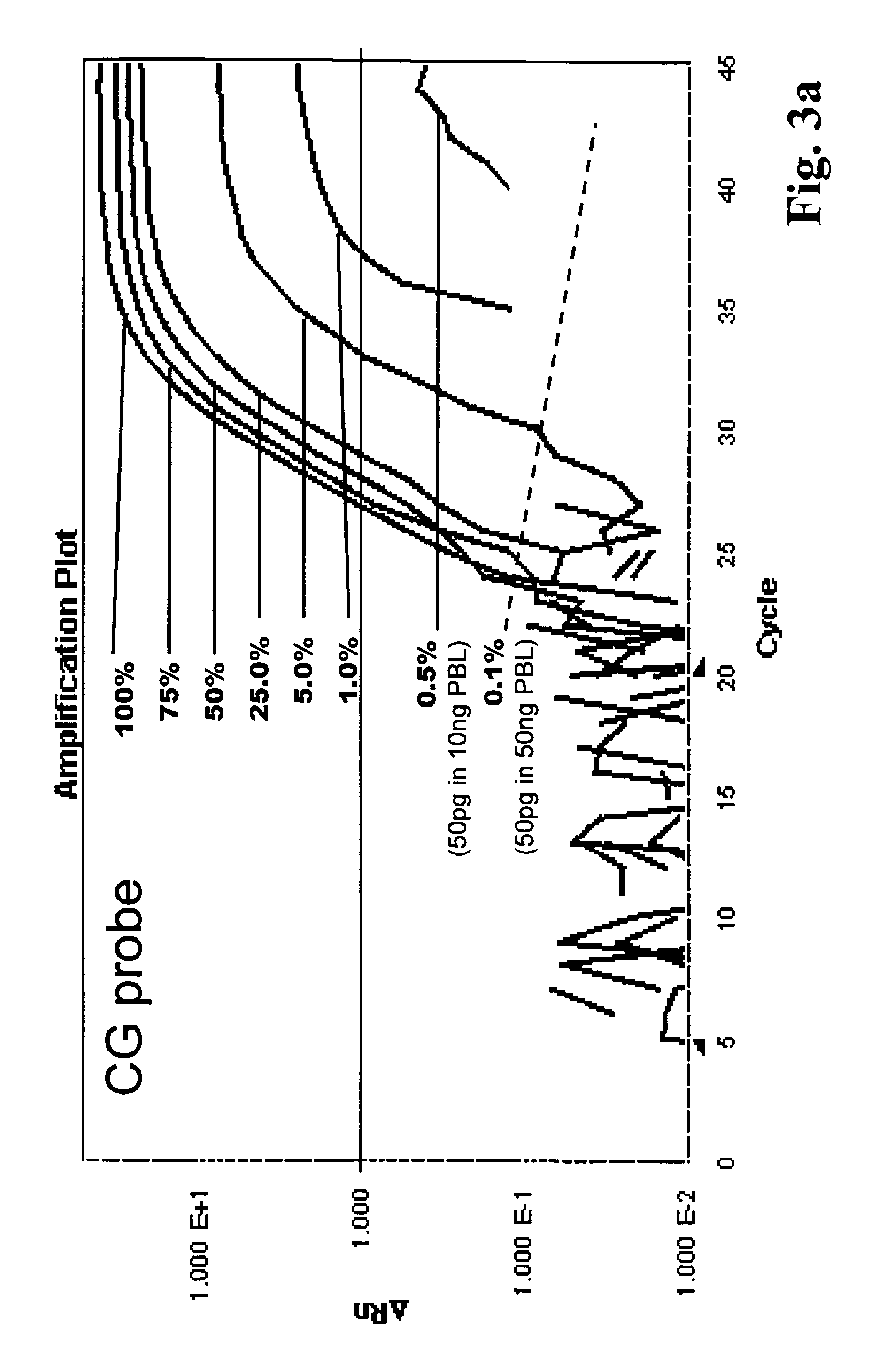
The present invention relates to a method for methylation analysis. It comprises the providing of a double stranded nucleic acid; its conversion, whereby unmethylated bases become distinguishable in their base-pairing behavior from methylated bases, and the analysis of both of the converted nucleic acid strands.
Owner:EPIGENOMICS AG
Method for methylation analysis of nucleic acid
ActiveUS8771939B2Sugar derivativesMicrobiological testing/measurementMethylation analysisAnalysis method
The present invention relates to a method for methylation analysis. It comprises the providing of a double stranded nucleic acid; its conversion, whereby unmethylated bases become distinguishable in their base-pairing behavior from methylated bases, and the analysis of both of the converted nucleic acid strands.
Owner:EPIGENOMICS AG
Branched α-glucan, α-glucosyltransferase which forms the glucan, their preparation and uses
ActiveUS8324375B2Easy to solveReducing lipidCosmetic preparationsOrganic active ingredientsAcetic acidDietary fiber
The present invention has objects to provide a glucan useful as water-soluble dietary fiber, its preparation and uses. The present invention solves the above objects by providing a branched α-glucan, which is constructed by glucose molecules and characterized by methylation analysis as follows:(1) Ratio of 2,3,6-trimethyl-1,4,5-triacetyl-glucitol to 2,3,4-trimethyl-1,5,6-triacetyl-glucitol is in the range of 1:0.6 to 1:4;(2) Total content of 2,3,6-trimethyl-1,4,5-triacetyl-glucitol and 2,3,4-trimethyl-1,5,6-triacetyl-glucitol is 60% or higher in the partially methylated glucitol acetates;(3) Content of 2,4,6-trimethyl-1,3,5-triacetyl-glucitol is 0.5% or higher but less than 10% in the partially methylated glucitol acetates; and(4) Content of 2,4-dimethyl-1,3,5,6-tetraacetyl-glucitol is 0.5% or higher in the partially methylated glucitol acetates; a novel α-glucosyltransferase which forms the branched α-glucan, processes for producing them, and their uses.
Owner:HAYASHIBARA BIOCHEMICAL LAB INC
Methylation analysis method and device for active region of circulating cell-free nucleosome, terminal equipment and storage medium
ActiveCN112735531AAuxiliary early diagnosisAid early screeningProteomicsGenomicsCell freeTerminal equipment
The invention provides a methylation analysis method and device for an active region of circulating cell-free nucleosome, terminal equipment and a storage medium, and the method comprises the steps: obtaining capture sequencing data of a to-be-detected plasma sample, and extracting cfDNA molecular fragments from the capture sequencing data; based on the extracted cfDNA molecular fragments, adopting windows to perform sliding operation in genome intervals of the cfDNA molecular fragments, and calculating the ratio of the number of cfDNA molecules crossing the whole window end to end in each window to the number of all cfDNA molecules in different conditions covered by the window; based on the calculated ratio, screening out an interval having significant difference with a baseline nucleosome activity difference area created according to the healthy person sample through a Kolmonov Schmidov test method to obtain a nucleosome activity area; calculating the methylation phenotypic characteristics of the screened nucleosome active area, completing methylation analysis of the circulating acellular nucleosome active area. The method can effectively assist in distinguishing the source of the plasma sample to be detected.
Owner:臻和(北京)生物科技有限公司 +1
Methods and compositions related to quantitative, array based methylation analysis
ActiveUS20110281746A1Nucleotide librariesMicrobiological testing/measurementMethylation analysisBioinformatics
Owner:UNIV OF UTAH RES FOUND
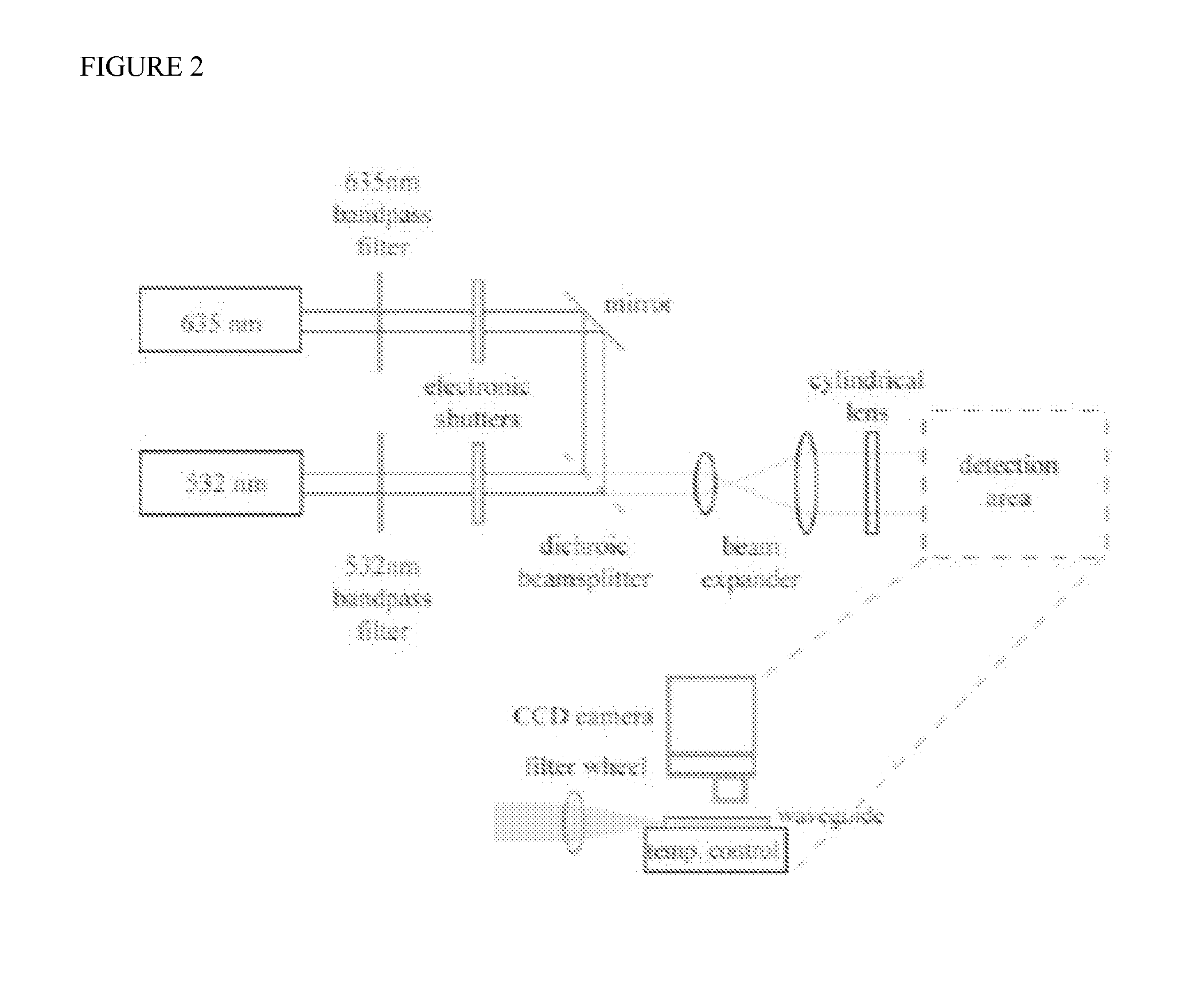
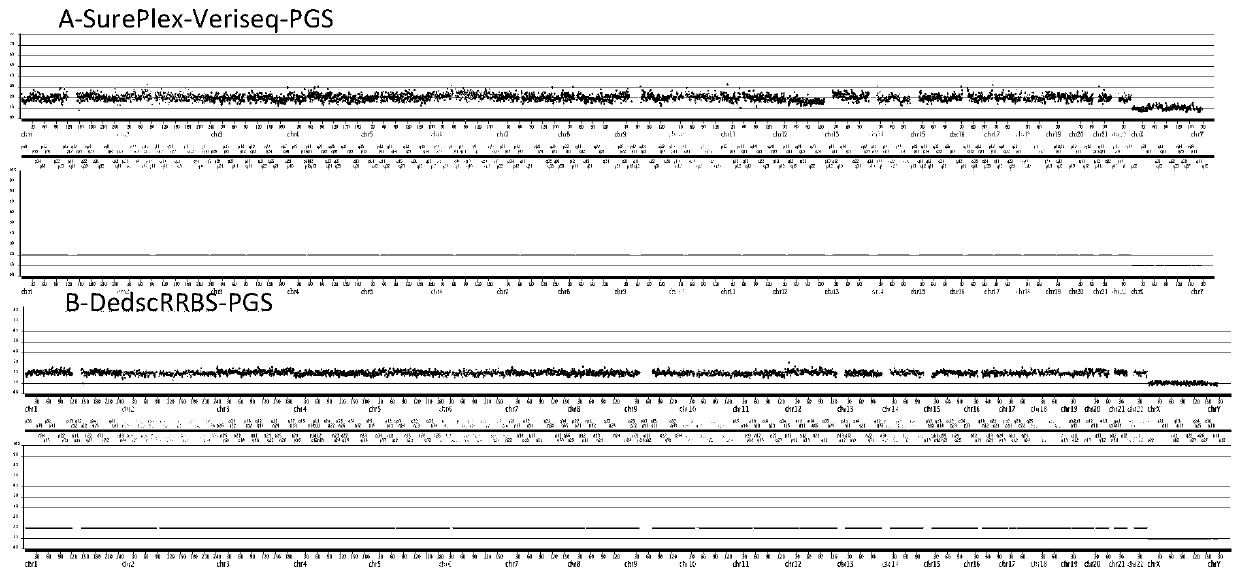
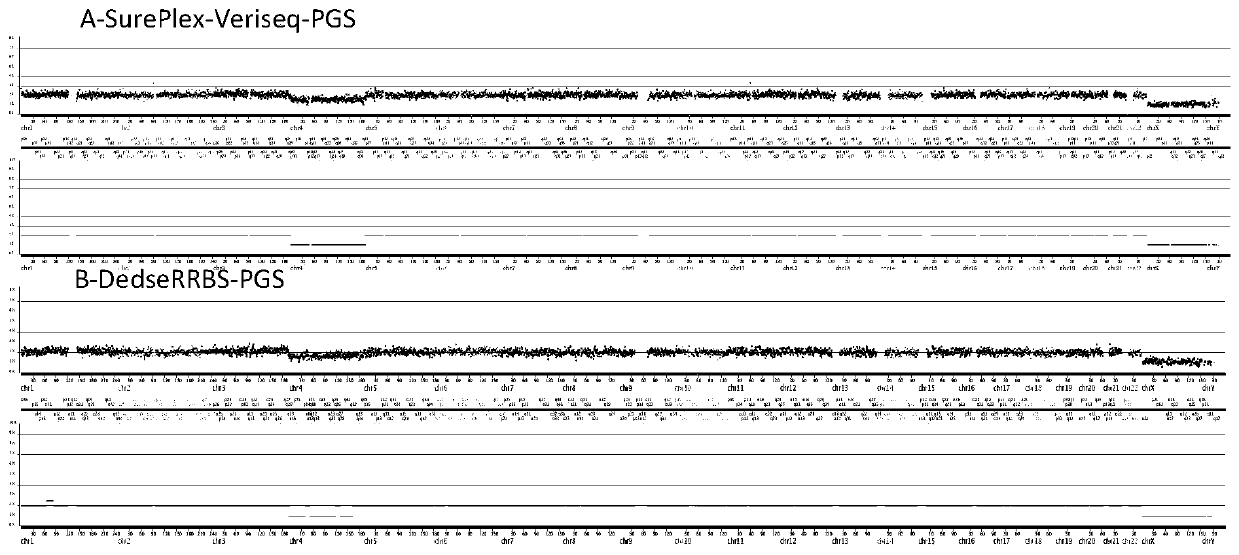
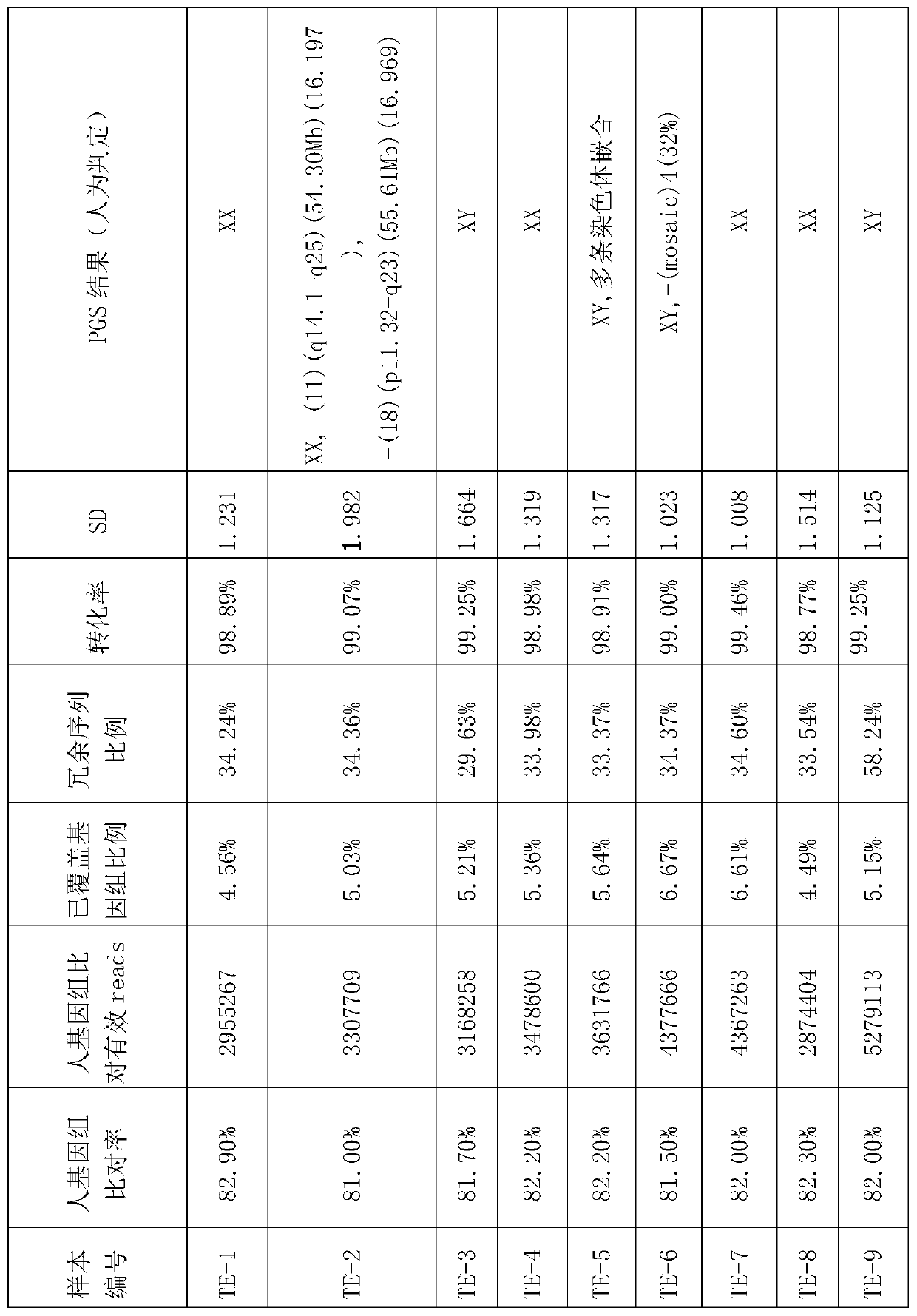
Method for conducting PGS and methylation analysis on biopsy cells by means of DedscRRBS analysis method
PendingCN109943628AIncrease success rateMicrobiological testing/measurementDNA methylationGerm layer
The invention relates to a method for parsing the blastocyst ectoderm cell DNA methylation status by means of a phenogenetics analysis technology by taking 'test tube baby' blastocyst ectoderm cells as raw materials and adopting a double-enzyme digestion single-cell reduced-representation bifulfite sequencing technology and conducting preimplantation genetic screening on the chromosome state. Thedevelopment potential of an embryo is comprehensively evaluated from various angles, a new reference is provided for selecting the 'right' embryo in assisted reproduction, and strong support is provided for increasing the success rate of the 'test-tube baby' cycle.
Owner:北京中科遗传与生殖医学研究院有限责任公司
Method for methylation analysis
Aspects of the invention relate to composition and methods for the providing of DNA for methylation analysis that is in particular suitable to be applied in reference laboratories. Further 5 aspects of the invention relate to composition and methods for the highly specific and sensitive methylation analysis of the Septin 9 gene also in particular suitable to be applied in reference laboratories.
Owner:EPIGENOMICS AG
Methods and compositions related to a multi-methylation assay to predict patient outcome
InactiveUS20110223180A1Favorable methylationImproved prognosisMicrobiological testing/measurementGenetic material ingredientsBrain tumorMethylation analysis
Methods and compositions for the prognosis and classification of cancer, especially brain tumor, are provided. For example, in certain aspects methods for cancer prognosis using methylation analysis of selected biomarkers are described.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Gene methylation analysis method and product and application
PendingCN110157775AReduce degradationReduce lossMicrobiological testing/measurementDNA methylationCytosine
The invention relates to a gene methylation analysis method and product and application and provides a DNA methylation analysis method. The DNA methylation analysis method comprises the steps that 1)a biologic sample with DNA is in contact with a carrier with a nucleic acid adsorption capacity and a lysis solution with a nucleic acid adsorption capacity at the same time, and the DNA is gathered on the carrier; 2) the carrier, adsorbing the DNA, obtained in the first step is in direct contact with a conversion reagent, and accordingly one or more non-methylated cytosine basic groups of the DNAgathered on the carrier are converted into cytosine or other basic groups, different from cytosine, detectable in hybridization; 3) a combination solution is used for treating a mixed solution obtained in the second step, so that the converted DNA is gathered on the carrier again. The invention further provides a corresponding gene methylation detection method, a kit and application.
Owner:JIANGSU MICRODIAG BIOMEDICINE TECH CO LTD
Methylation analysis on self-samples as triage tool for hpv-positive women
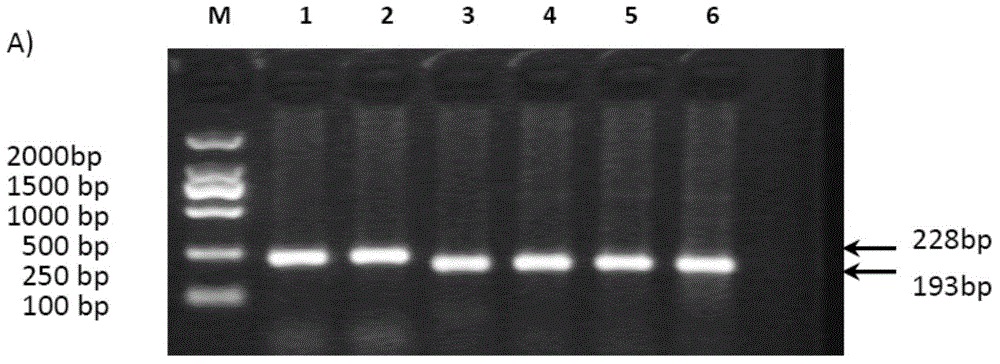
The inventors now have developed a (molecular) diagnostic assay based on detection of alterations in markers MAL and hsa-mi R124, in particular promoter hypermethylation, to identify human papillomavirus (HPV)-induced high-grade precancerous lesions such as premalignant cervical lesions of invasive cervical cancer, within cell material obtained via self-sampling. In particular, the present invention relates to the use of the MAL gene (including its promoter) and the hsa-mi R124 gene as marker for HPV-induced high-grade premalignant lesions, allowing early detection and a better treatment option for the individual patient.
Owner:SELF SCREEN
Method for identifying imprinted gene Rasgrf1 of domestic pig
Owner:ANHUI AGRICULTURAL UNIVERSITY
Methylation Analysis On Self-samples As Triage Tool For HPV-positive Women
The inventors now have developed a (molecular) diagnostic assay based on detection of alterations in markers MAL and hsa-mi R124, in particular promoter hypermethylation, to identify human papillomavirus (HPV)-induced high-grade precancerous lesions such as premalignant cervical lesions of invasive cervical cancer, within cell material obtained via self- sampling. In particular, the present invention relates to the use of the MAL gene (including its promoter) and the hsa-mi R124 gene as marker for HPV- induced high-grade premalignant lesions, allowing early detection and a better treatment option for the individual patient.
Owner:SELF SCREEN
Control nucleic acid constructs for use in analysis of methylation status
InactiveUS20080102452A1ConfidenceSugar derivativesMicrobiological testing/measurementNucleotideMethylation analysis
In some embodiments, control nucleic acid constructs useful as spiking reagents are provided which comprise a nucleic acid vector having an insert comprising a control nucleic acid molecule. In some embodiments, the insert contains at least one methyltransferase recognition site, such as a CpG dinucleotide. In some embodiments, the insert has a sequence complementary to a negative control probe of a microarray. Methods and kits for using the control nucleic acid constructs as spiking reagents in methylation analysis are disclosed.
Owner:AGILENT TECH INC
Preparation of templates for methylation analysis
ActiveUS9868982B2Convenient treatmentGood orientationMicrobiological testing/measurementCytosineNucleotide
The invention relates to a method of preparing and using a library of template polynucleotides suitable for use as templates in solid-phase nucleic acid amplification and sequencing reactions to determine the methylation status of the cytosine bases in the library. In particular, the invention relates to a method of preparing and analyzing a library of template polynucleotides suitable for methylation analysis.
Owner:MASSACHUSETTS INST OF TECH +1
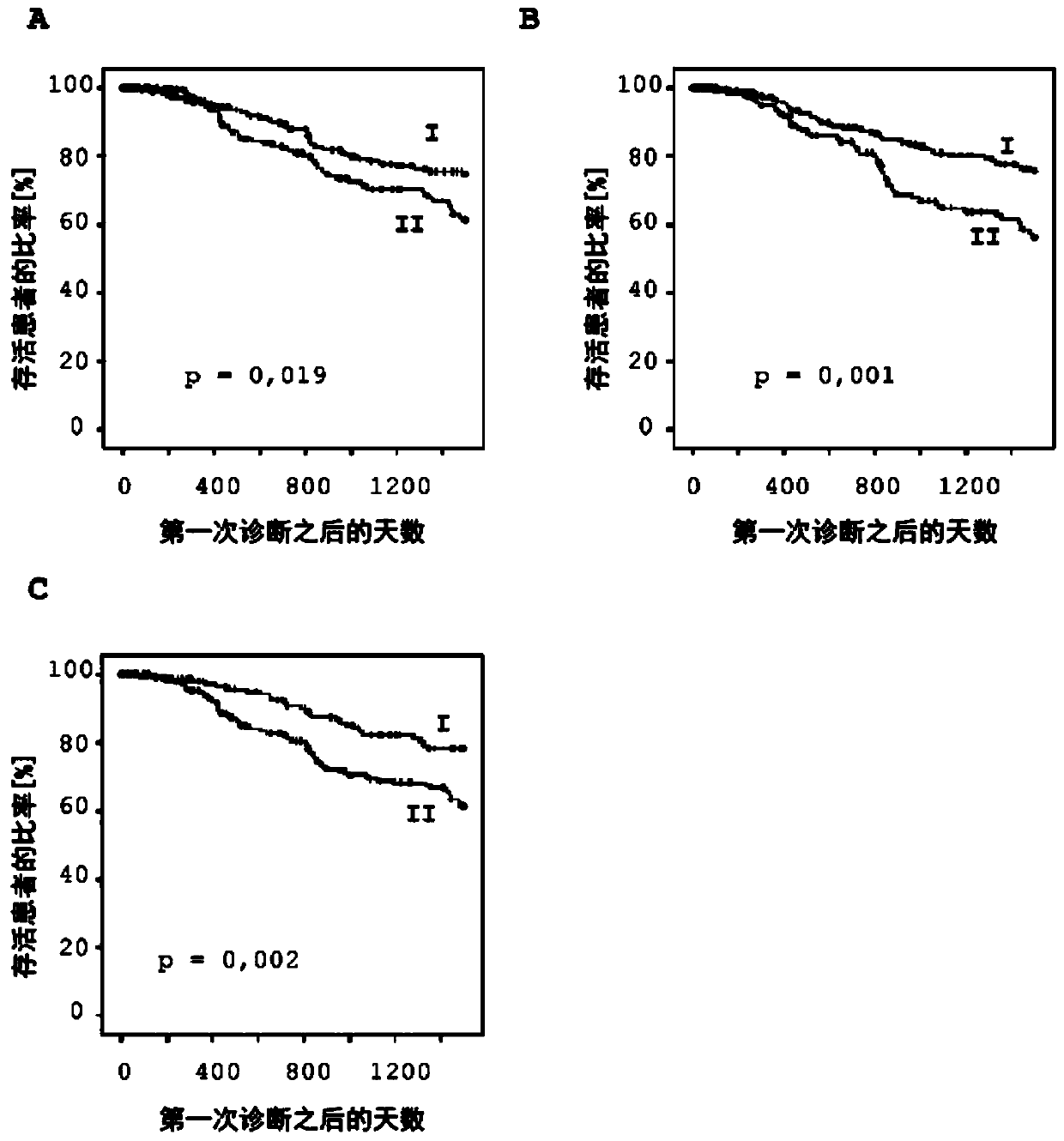
Method for determining the response of a malignant disease to an immunotherapy
PendingCN111630184AReliable predictionMicrobiological testing/measurementAntibody medical ingredientsDiseaseCD86
The invention relates to a method for predicting the response of a malignant disease to an immunotherapy which inhibits the PD-1 immune checkpoint signal path. The method is characterized in that at least one part of an immuno-regulatory gene selected from CTLA4, CD86, CD28, CD80 and / or ICOS of cells of the malignant disease and / or immune cells which interact with the cells of the malignant disease is subjected to a DNA methylation analysis, and the response of the malignant disease to the immunotherapy is predicted using the result.
Owner:迪莫迪特里希
DNA methylation marker for predicting risk of primary breast cancer, and screening method and application of DNA methylation marker
PendingCN111440869AImprove the detection rateLow costMicrobiological testing/measurementBiostatisticsDNA methylationPrimary breast cancer
The invention discloses a DNA methylation marker for predicting the risk of primary breast cancer, and a screening method and application of the DNA methylation marker. The method comprises the following steps: (1) performing methylation analysis on sample data to obtain methylation sites related to prediction of primary breast cancer occurrence risks; (2) obtaining methylation Beta values for themethylation sites; (3) constructing a primary breast cancer occurrence risk prediction model based on the methylation Beta values of the methylation sites, and verifying the feasibility of the modelby calculating a ratio; (4) constructing a primary breast cancer occurrence risk prediction model based on the methylation sites by adopting a machine learning method, calculating a ratio ratio, AUC,a recall rate, an accuracy rate and an F1 value, and performing mutual verification with the prediction model in the step (3); and (5) obtaining the methylation sites corresponding to the methylationprobe in the prediction model as the DNA methylation marker. The DNA methylation marker can improve the detection rate of primary breast cancer, and is suitable for large-scale popularization and application.
Owner:武汉百药联科科技有限公司 +1
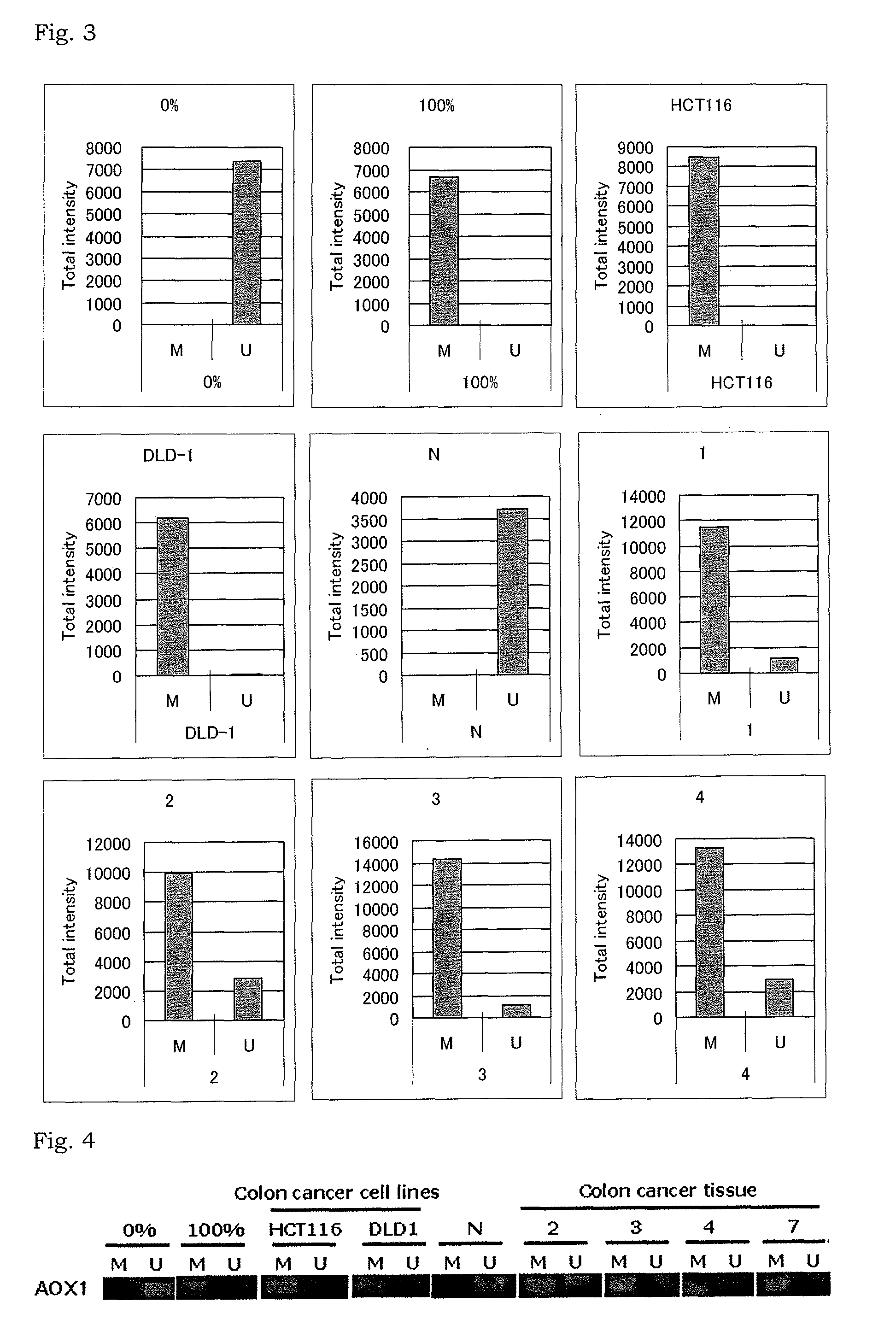
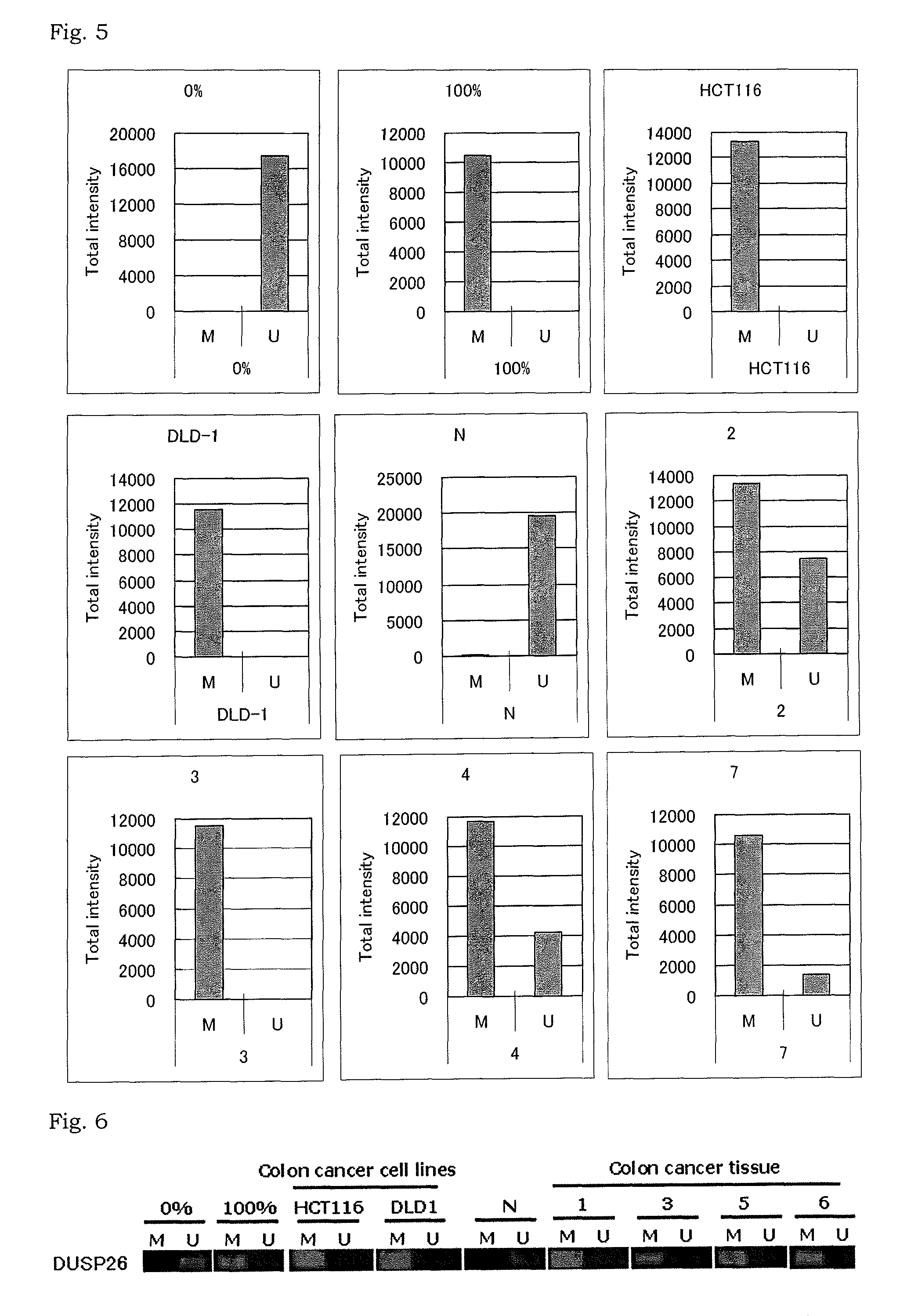
Methylation analysis to determine the presence of cancer cells
InactiveUS9075063B2Sugar derivativesMicrobiological testing/measurementCancer cellMethylation analysis
The present invention relates to a method for determination of the presence or absence of cancer cells in a biological sample or a method for determination of the prognosis of a colorectal cancer patient based on a result obtained by extracting DNA from a biological sample and analyzing methylation status of a marker gene in the DNA.
Owner:THE UNIV OF TOKYO +1
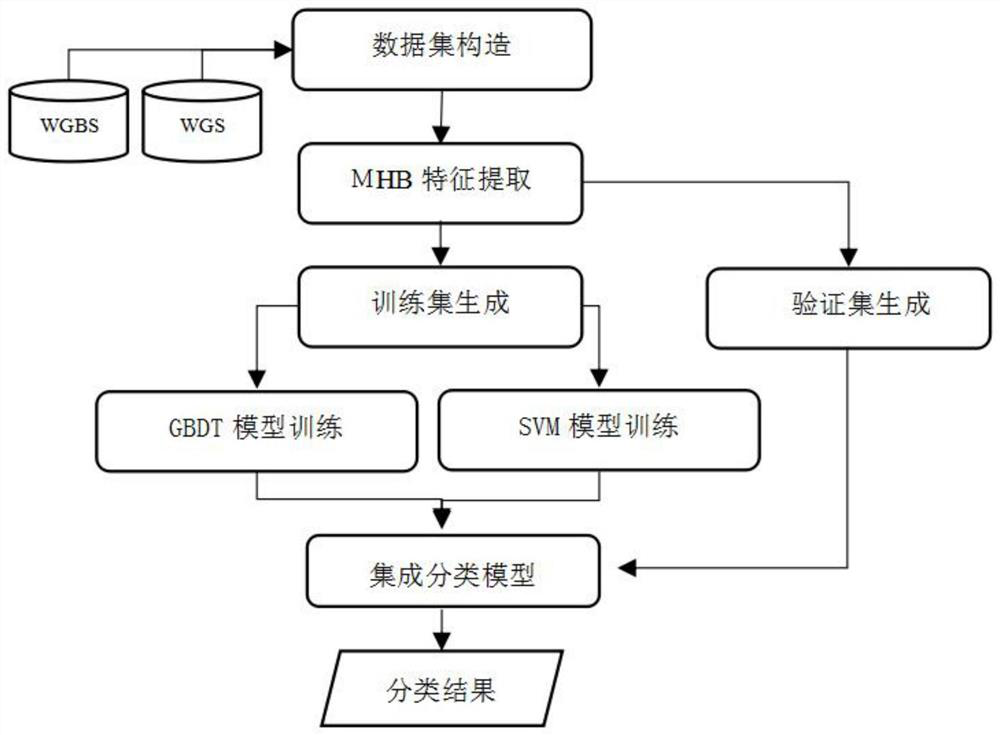
Plasma sample cancer early screening method based on ensemble learning
InactiveCN113611404AImprove performanceImprove stabilityMicrobiological testing/measurementBiostatisticsScreening methodEngineering
The invention discloses a plasma sample cancer early screening method based on ensemble learning, and belongs to the field of cancer early screening. The cancer early screening method comprises the following steps: 1, taking data obtained by performing characteristic value extraction on ctDNA mutation and methylation analysis data in plasma as a training set and a verification set, and then respectively importing the training set into a gradient boosting tree model and a classification model of a support vector machine; 2, fusing the gradient boosting tree model trained in the step 1 and the classification model of the support vector machine trained in the step 1 to obtain an integrated classification model; 3, importing the verification set in the step 1 into the integrated classification model in the step 3, and obtaining a classification result through a voting mechanism, namely, a cancer early screening result. The performance of the model is optimized under different training conditions, the adaptability to sample size, sample feature distribution and the like during model training is enhanced, the stability of the model is effectively improved, the reliability in practical application is ensured, and stable prediction precision is generated.
Owner:哈尔滨智吾康科技有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com