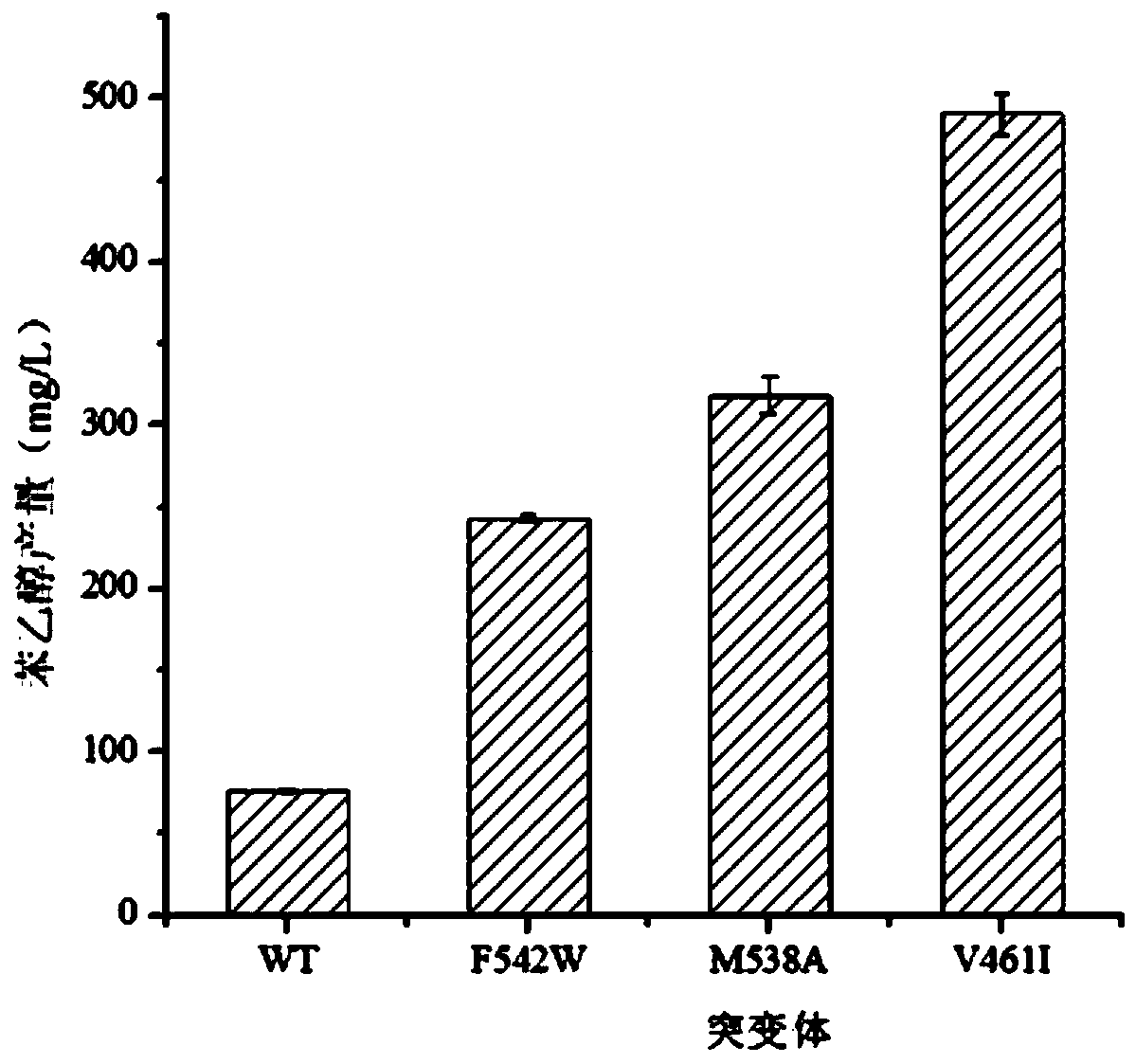
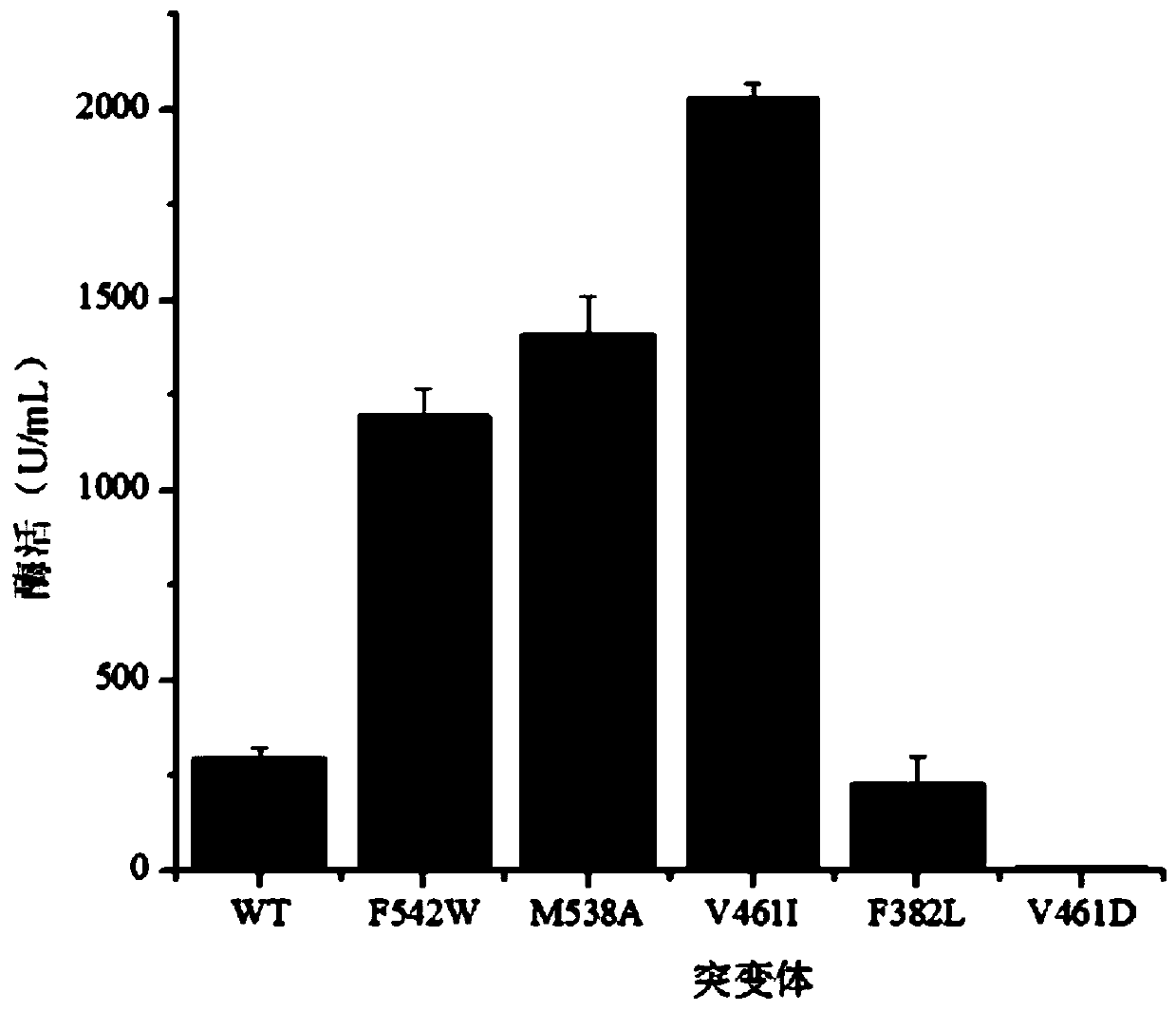
Application of phenylpyruvate decarboxylase mutant M538A in biological fermentation production of phenethyl alcohol
A technology of phenylpyruvate decarboxylase and biological fermentation, which is applied in the field of enzyme engineering and genetic engineering, can solve the problems of low catalytic efficiency and low substrate specificity, and achieve the goal of improving catalytic efficiency, improving enzyme production ability, and improving enzyme activity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Construction of recombinant plasmid pHY-P43-kivD:
[0022] An α-ketoacid decarboxylase kivD gene was cloned from Lactococcus lactis subsp.lactis (CICC6246) by using KivD-F and KivD-R. Using the Bacillus subtilis 168 genome as a template, the P43 promoter was amplified with P43-F and P43-R primers; the amylase terminator TamyL was amplified with TamyL-F and TamyL-R primers. P43-F and TamyL-R were used as primers to perform SOE-PCR on the three fragments of P43, kivD and TamyL to obtain the fusion fragment P43-kivD-TamyL. Using the plasmid pHY300PLK as a template and pHY-T5-F and pHY-T5-R as primers, the whole plasmid was amplified by PCR to obtain a linearized pHY300PLK vector. After the above amplified products were checked by electrophoresis, the PCR products were purified and recovered using a gel recovery kit. Using the ClonExpress II one-step cloning kit, the fusion fragment was fused with the linearized vector pHY300PLK to obtain the recombinant plasmid pHY-P43-k...
Embodiment 2
[0033] Preparation and enzyme activity determination of phenylpyruvate decarboxylase wild bacteria (WT):
[0034] The plasmid pHY-P43-kivD sequenced correctly in Example 1 was transformed into Bacillus licheniformis DW2. Select the transformant and inoculate it into LB medium after verifying that it is correct, culture it at 37°C for 14 hours; transfer it to the fermentation medium with an inoculum of 3%, culture it at 37°C for 24 hours, collect the bacteria by centrifugation, wash the bacteria twice with PBS, Finally, resuspend the cells with 1 ml of 50 mM potassium phosphate buffer (pH 6.8). Cells were disrupted using an ultrasonic breaker. Ultrasonic settings: 150W, 20kHz, working for 2s; off for 2s, a total of 8 minutes, centrifuged at 4°C to collect the supernatant, which was the crude enzyme solution.
[0035] The supernatant was tested for enzyme activity according to the following system, enzymatic reaction system (200μL): 50mM potassium phosphate buffer (pH 6.8), 1mM...
Embodiment 3
[0037] Preparation of Phenylpyruvate Decarboxylase Mutants
[0038] Using the constructed pHY-P43-kivD as a template, primers were designed for PCR amplification of the whole plasmid to obtain a linearized pHY300PLK vector with P43 promoter and amylase TamyL terminator. Perform site-directed mutation on the catalytic active site of kivD, and replace the mutation on the gene sequence by means of primers, specifically:
[0039] 1) Mutate the valine at position 461 of the catalytic domain of the phenylpyruvate decarboxylase molecule to isoleucine, split the base GTC encoding the amino acid at position 461 of kivD into two parts, primers V461I-AR and V461I -BF, using the plasmid in Example 1 as a template to amplify the upper and lower sections of the kivd mutant sequence; then use kivD-F and kivD-R as primers to perform SOE-PCR on the two sections to amplify the kivD mutant sequence V461I;
[0040] 2) Mutate the 538th methionine to alanine, split the base GCG encoding the 538th ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com