Base editing tools and their applications and methods for wide-window and sequence-bias-free base editing in eukaryotic cells
A technology of base editing and tools, applied in the field of base editing, which can solve problems such as limited applications
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0040] According to the present invention, the sequence of the linker may be any amino acid sequence capable of connecting the amino acid sequences at both ends, and the present invention has no particular limitation on this. According to a preferred embodiment of the present invention, the linker The amino acid sequence of is shown in SEQ ID NO:1.
[0041]According to the present invention, the cytosine deaminase AID can be any amino acid sequence having cytosine deaminase AID activity known in the art, and according to a preferred embodiment of the present invention, the cytosine deaminase is The amino acid sequence of the full-length AIDfl is shown in SEQ ID NO:4.
[0042] However, the inventors of the present invention found in research that by performing point mutation and C-terminal truncation on the amino acid sequence shown in SEQ ID NO: 4, the base editing efficiency of the final base editing tool can be effectively improved, Wherein, the point mutation is selected f...
Embodiment 1
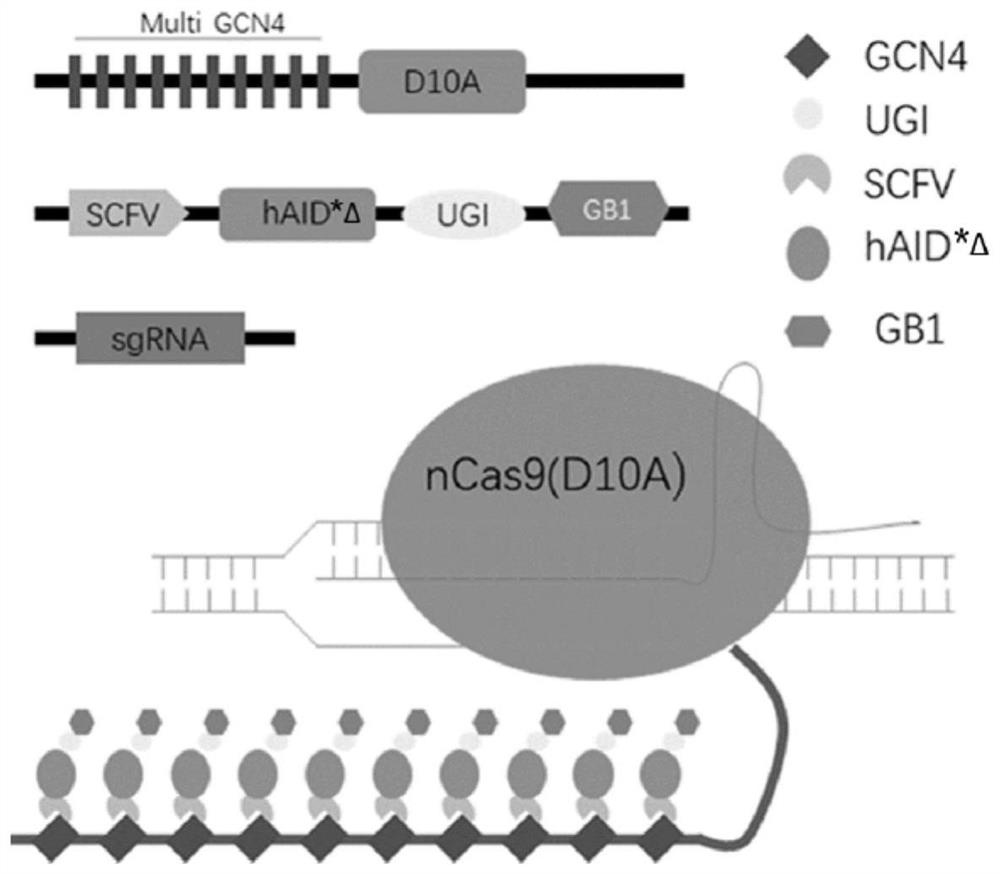
[0066] This example is used to illustrate the construction of a plasmid encoding scFv-AID-UGI-GB1 protein
[0067] 1. Amplification of nucleic acid sequence fragment (AID) encoding AID
[0068] The coding sequence hAID*Δ of the optimized version of cytosine deaminase AID hAID*Δ synthesized by Jinweizhi Biotechnology Co., Ltd. (the encoded amino acid sequence is shown in SEQ ID NO: 5, and the DNA sequence is shown in SEQ ID NO: 139) , cloned in the pUC57 vector to obtain the pUC57-hAID*Δ vector, diluted to 10 μM as a PCR template. Design forward primer (as shown in SEQ ID NO: 10), reverse primer (as shown in SEQ ID NO: 11), add water and dissolve to 10 μM. The hAID sequence fragment was amplified using Novizan High Fidelity Enzyme Kit (Vazyme, p501-d2). The amplification system is shown in Table 1, and the PCR reaction conditions are shown in Table 2.
[0069] Table 1
[0070]
[0071] Table 2
[0072]
[0073] 2. Amplification of the nucleic acid sequence fragment (...
Embodiment 2
[0087] This example is used to illustrate the construction of the plasmid encoding scFv-AIDfl-UGI-GB1 protein
[0088] The construction of the plasmid encoding the scFv-AIDfl-UGI-GB1 protein was carried out according to Example 1, except that the human codon-optimized hAID*Δ was replaced with the nucleotide sequence of the unoptimized full-length AIDfl, that is, the encoding For the nucleotide sequence (SEQ ID NO: 138) of the protein shown in SEQ ID NO: 4, the amplification forward primer and reverse primer are shown in SEQ ID NO: 143 and SEQ ID NO: 144, respectively, to obtain BE-PA2.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com