Multiple enrichment method for detection of low-frequency mutations associated with targeted drugs in non-small cell lung cancer
A non-small cell lung cancer, low-frequency mutation technology, applied in biochemical equipment and methods, microbial measurement/testing, recombinant DNA technology, etc., can solve problems affecting detection specificity, complex data processing, long detection time, etc., to achieve Easy operation, easy and fast operation, high sensitivity effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
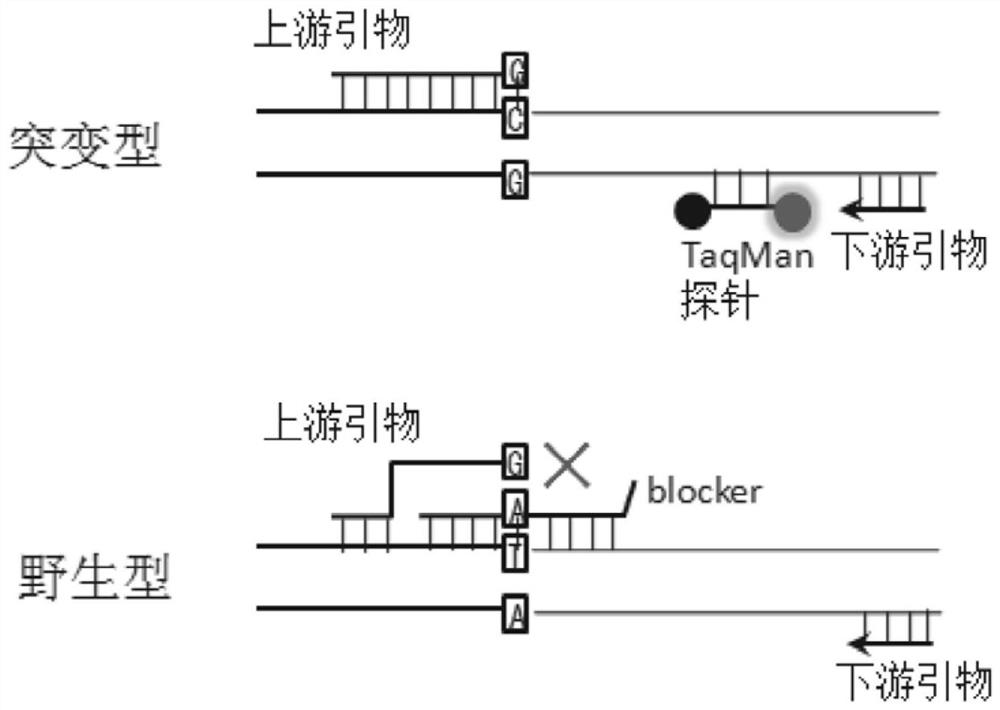
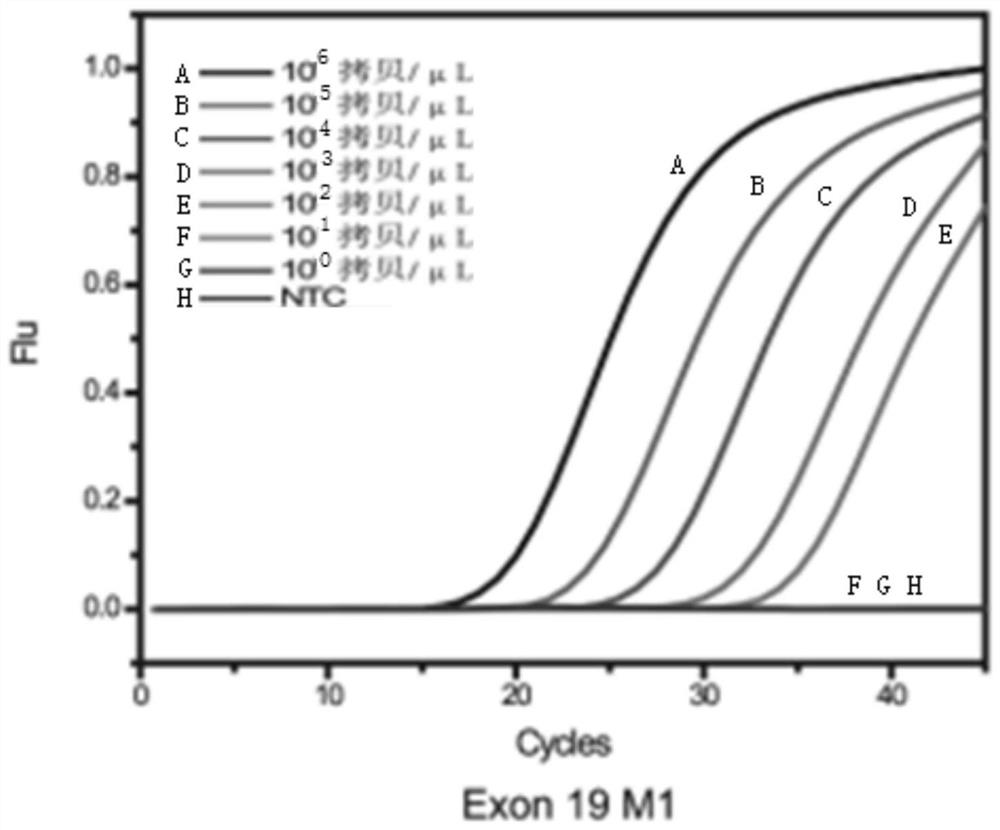
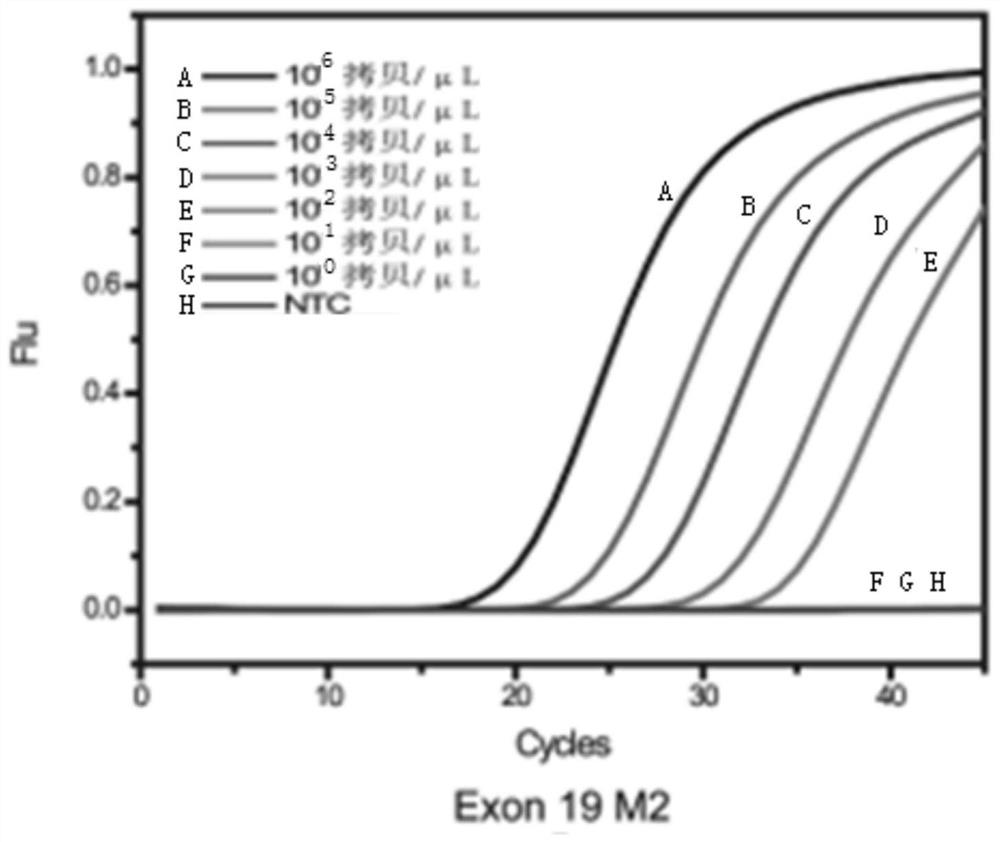
[0057] Example 1. Non-small cell lung cancer targeted drug-related low-frequency mutations 2235_2249del-15, 2236_2250del-15, T790M and L858R multiple enrichment detection method (TaqMan probe method)
[0058] 1. The composition of non-small cell lung cancer targeted drug-related low-frequency mutations 2235_2249del-15, 2236_2250del-15, T790M and L858R multiple enrichment detection kit (TaqMan probe method)
[0059] (1) There are four segments in the target sequence. Including: a section on exon 19 of the human EGFR gene, including the deletion mutation 2235_2249del-15 and 2236_2250del-15 and a total of 85 bp DNA sequence before and after; a section on exon 20 of the human EGFR gene, including the point mutation to be tested T790M and a total of 93bp DNA sequence before and after; a section on exon 21 of the human EGFR gene, including the point mutation L858R to be tested and a total of 109bp DNA sequence before and after; a section on the human β-actin gene, a total of 68bp. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com