Method for identifying controllable gene based on complex network structure
A technology of network structure and identification method, applied in the field of bioinformatics, which can solve the problems of information limitation, ignoring the interaction of neighbor nodes of nodes, complex network incompatibility, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0083] The present invention will be described in detail below in conjunction with the accompanying drawings and specific embodiments.
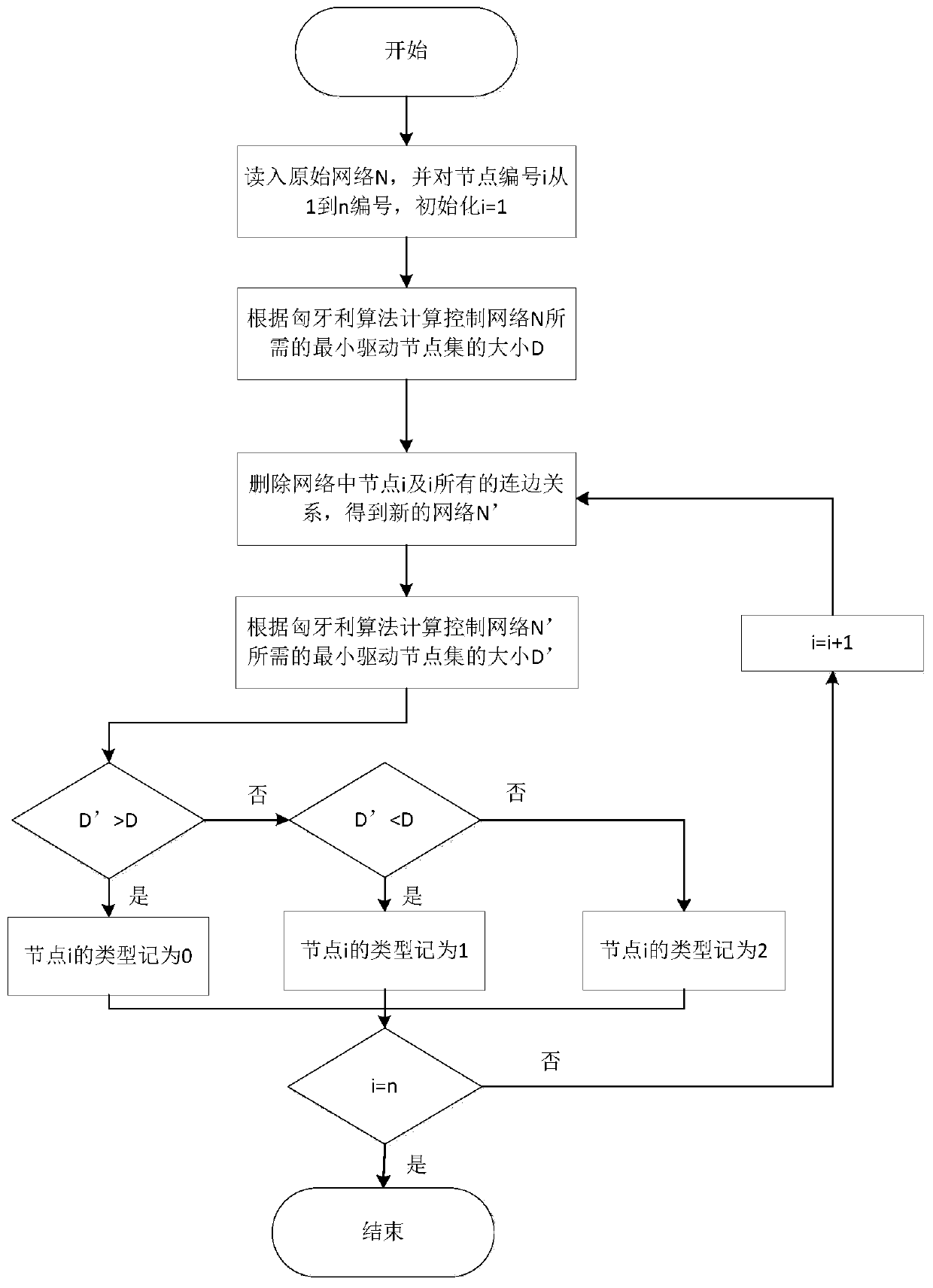
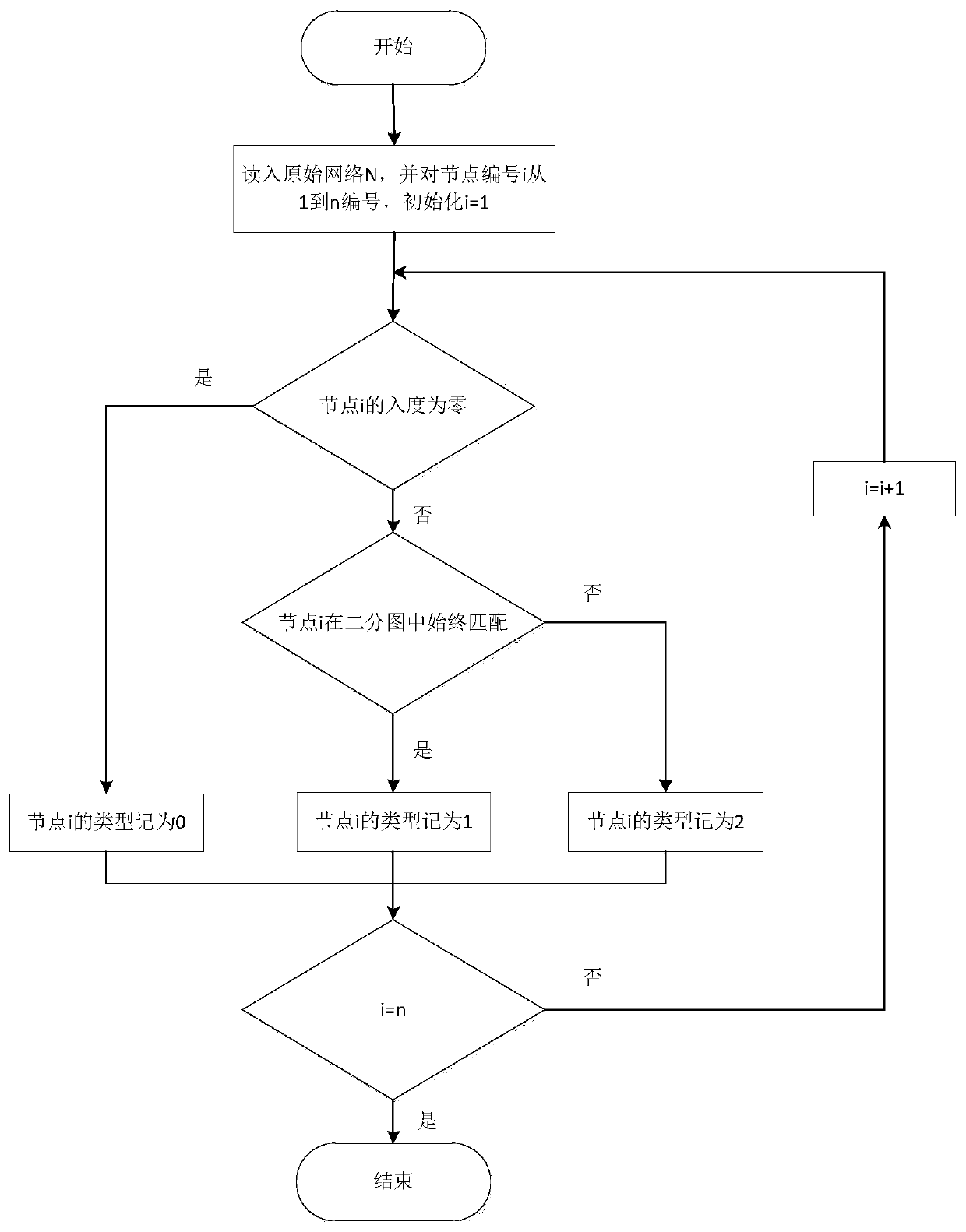
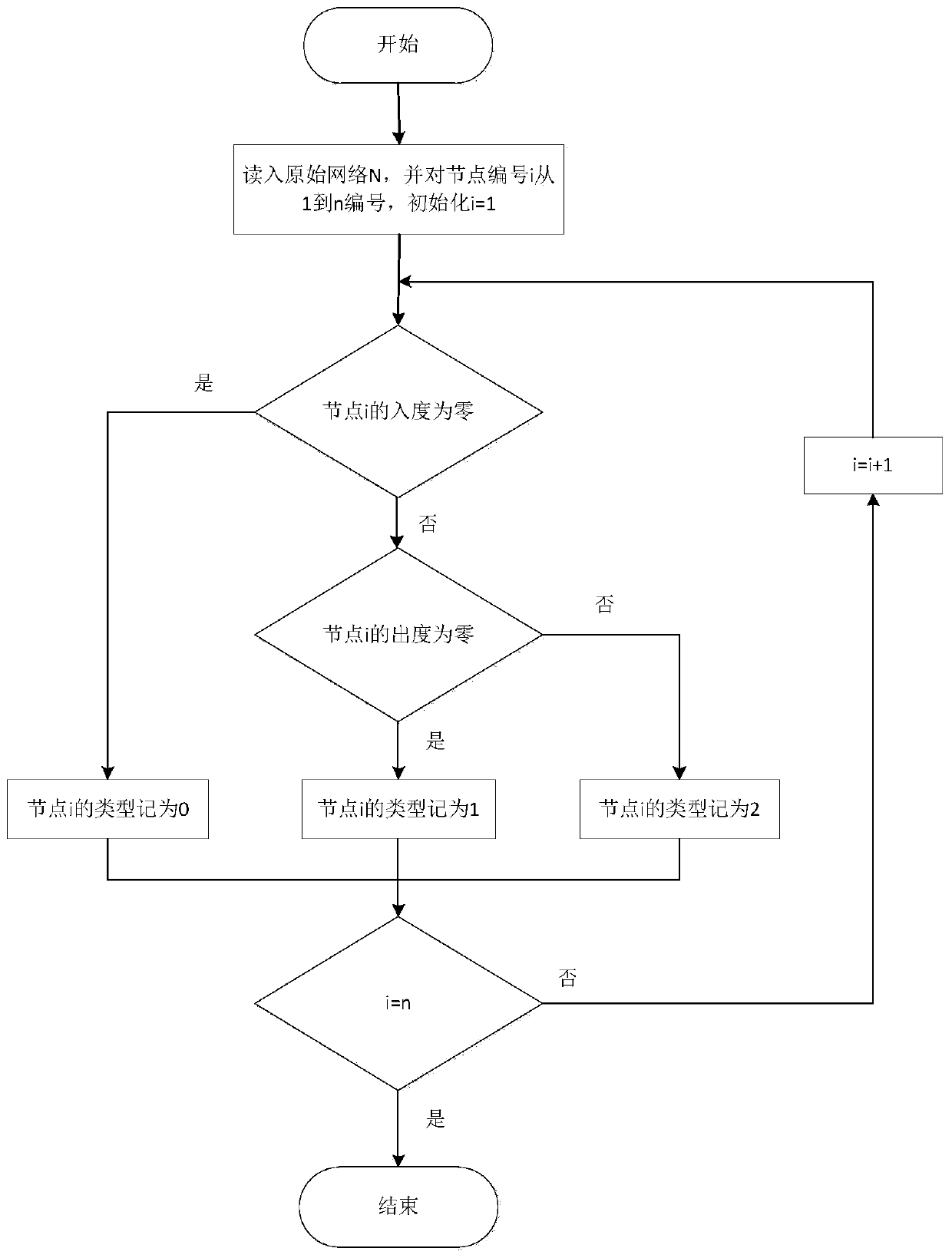
[0084] A controllable gene identification method based on complex network structure, such as Figure 5 shown, including the following steps:
[0085] Step 1: Reading data from tissue-specific regulatory networks
[0086] Each row in the original data contains two columns of data, representing the regulated gene and the regulated gene respectively;
[0087] Step 2: Classify the nodes in the network using four different node classification methods
[0088] The first is a node classification method based on controllability;
[0089] The second is a node classification method based on control capability;
[0090] The third is the node classification method based on the source of the control function;
[0091] The fourth is a node classification method based on the robustness of control edges;
[0092] Step 3: Combine the four classification...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com