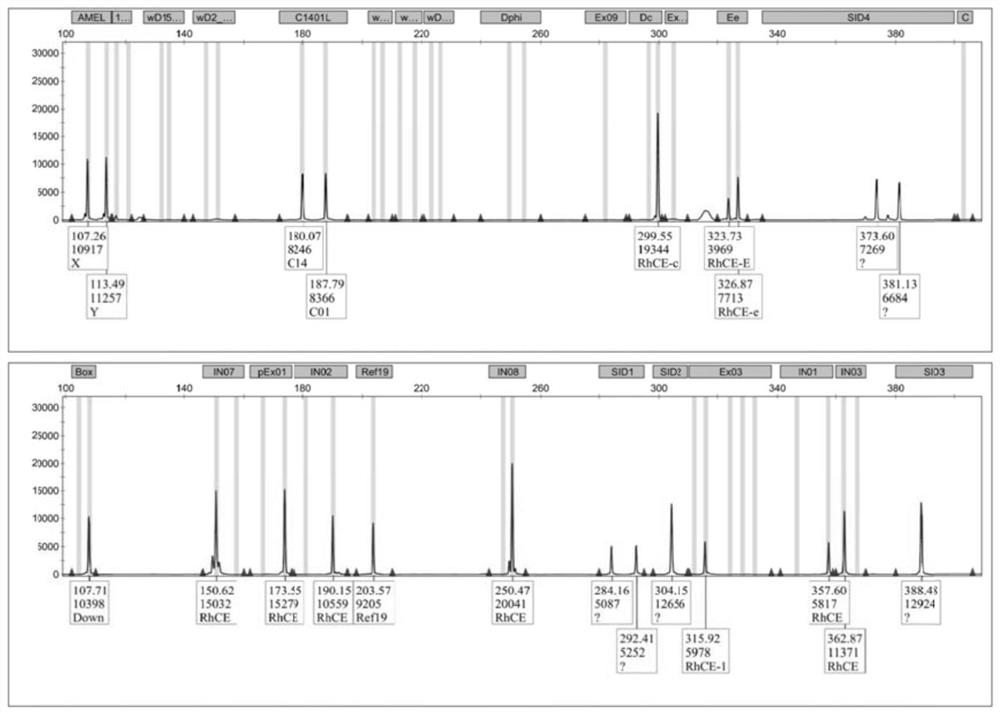
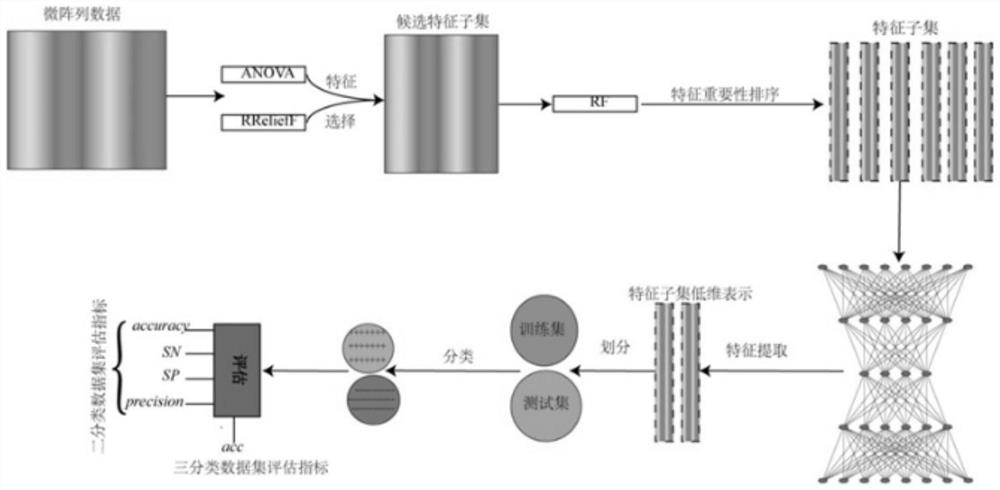
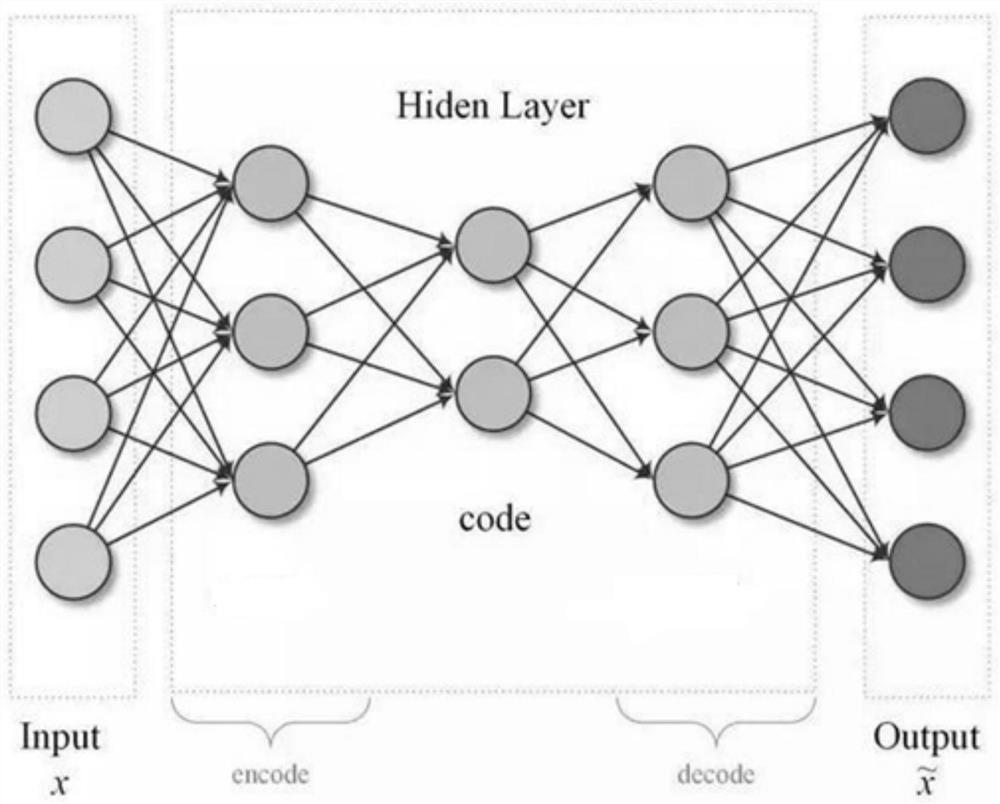
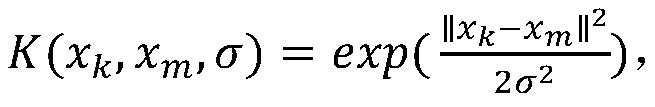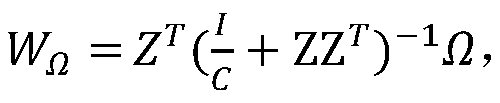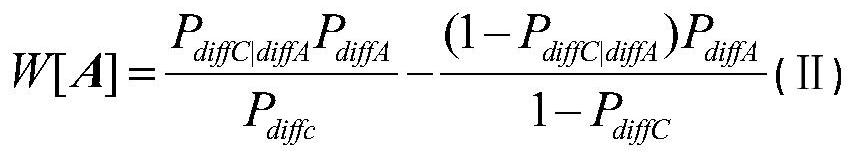Patents
Literature
39 results about "Gene classification" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
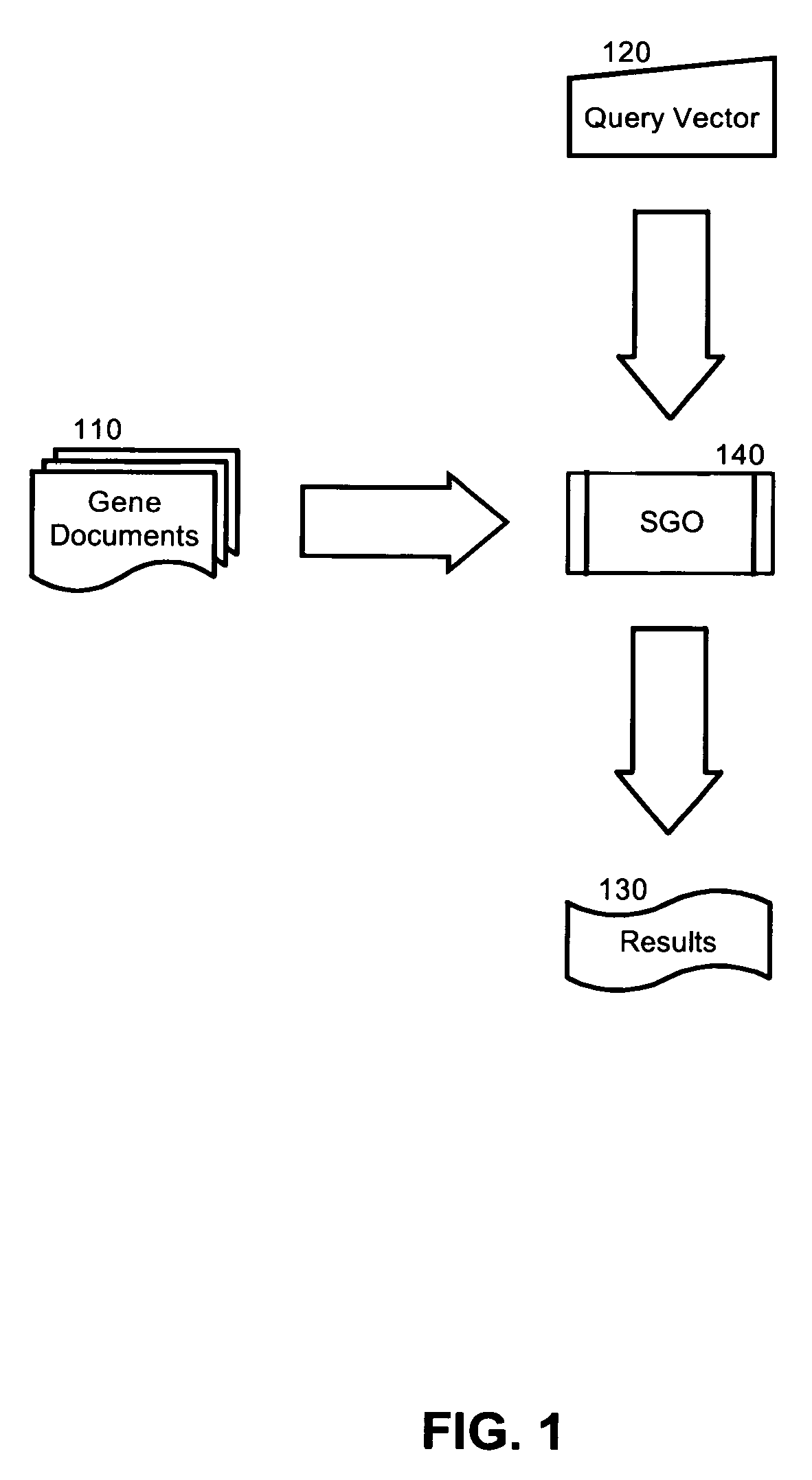
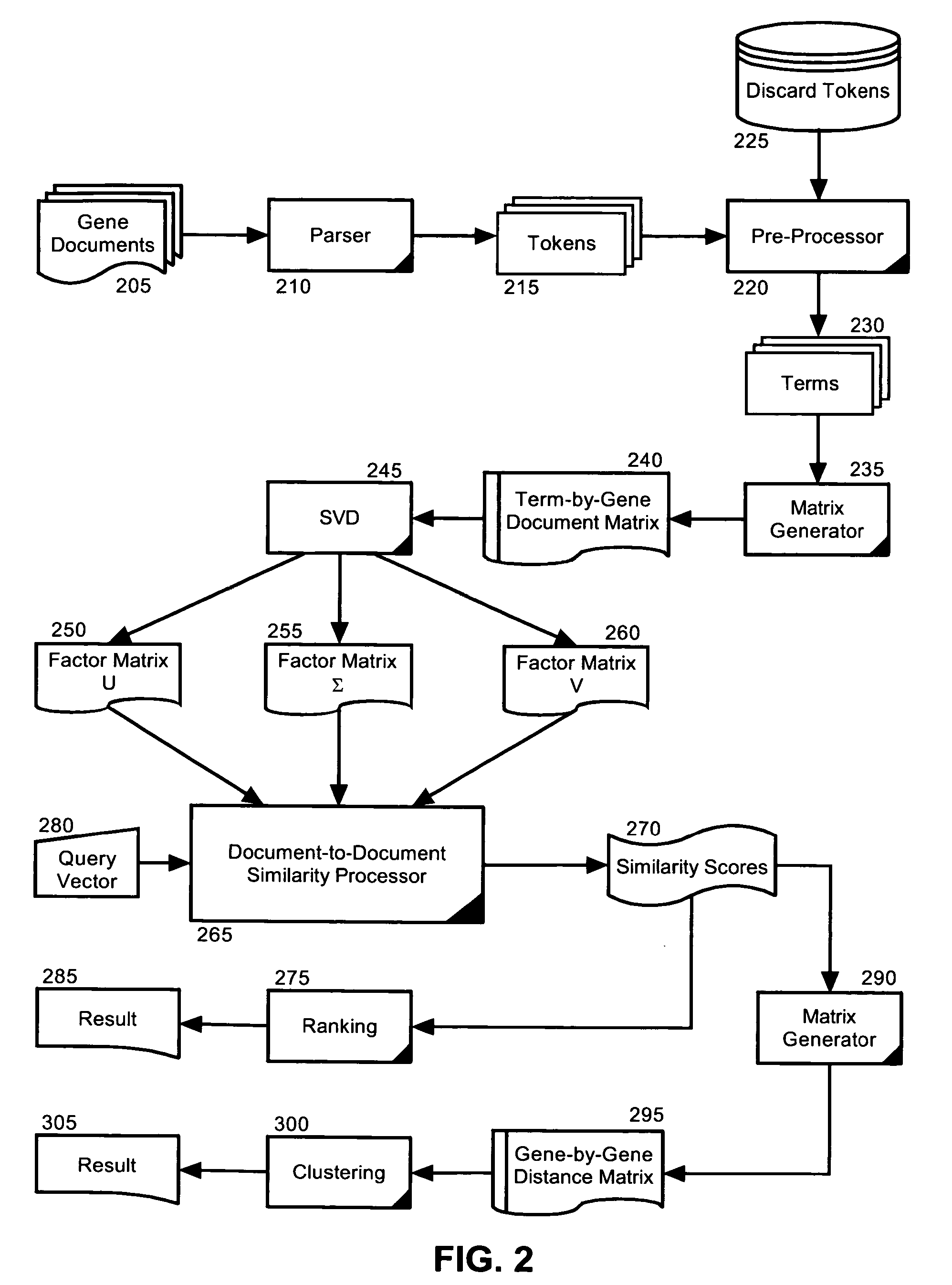
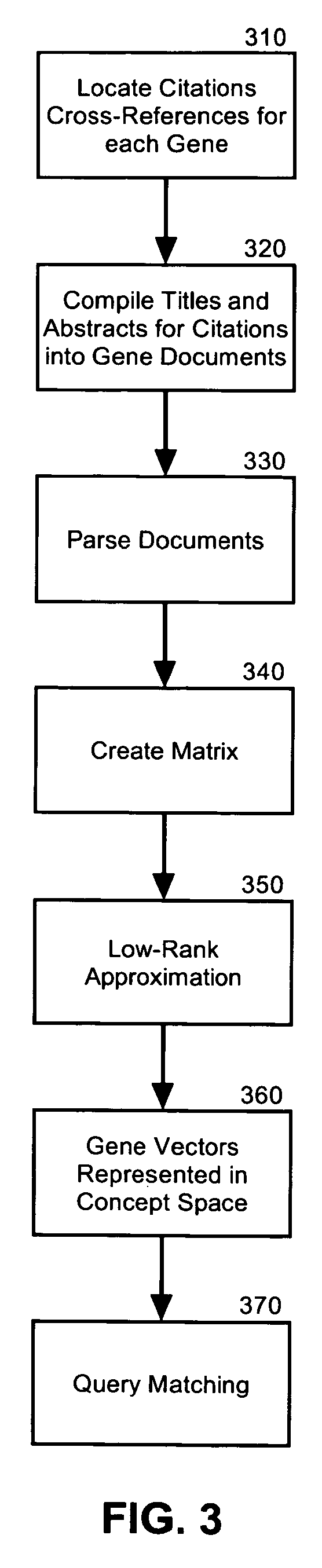
Semantic gene organizer
InactiveUS20060047441A1Rapidly and accurately classifyBiological testingSpecial data processing applicationsPaper documentDocument preparation
A semantic gene classification and annotation system, method and computer program can utilize Latent Semantic Indexing (LSI) to identify conceptually related genes based on textual information in biomedical literature, including MEDLINE citations. In addition, term weights calculated from the usage of the gene terms in and across gene documents can be used to automatically assign gene aliases and extend gene function annotation based upon primary biomedical literature.
Owner:UNIV OF TENNESSEE RES FOUND
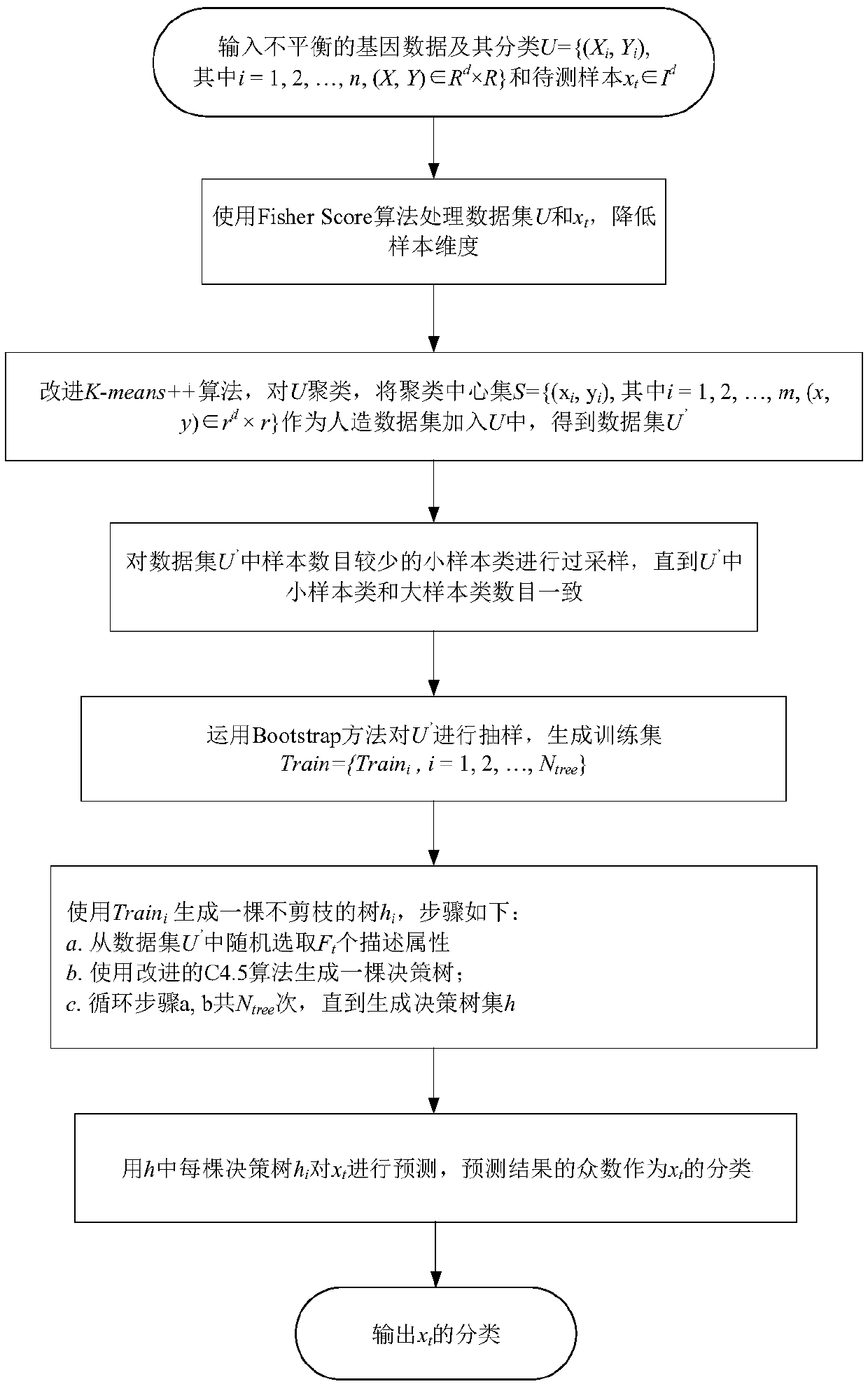
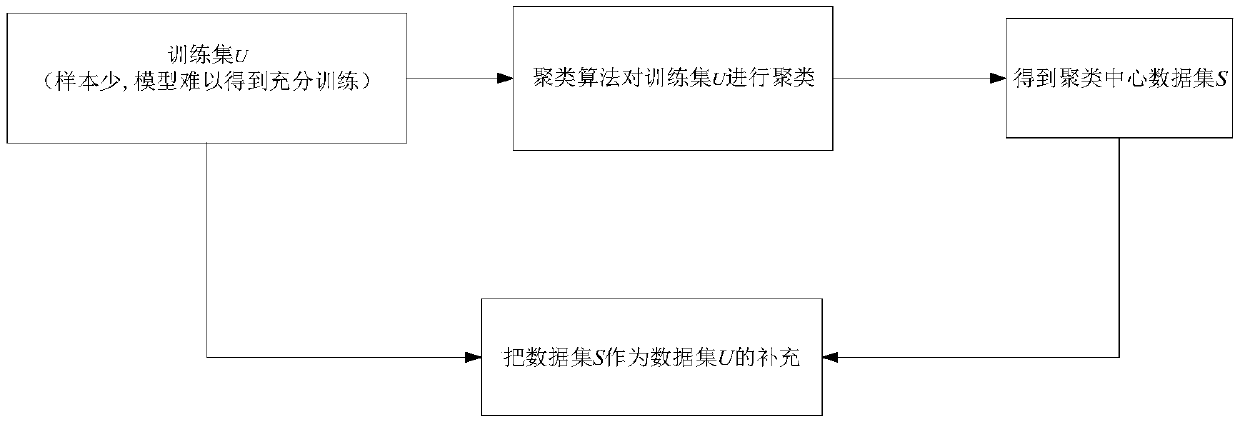
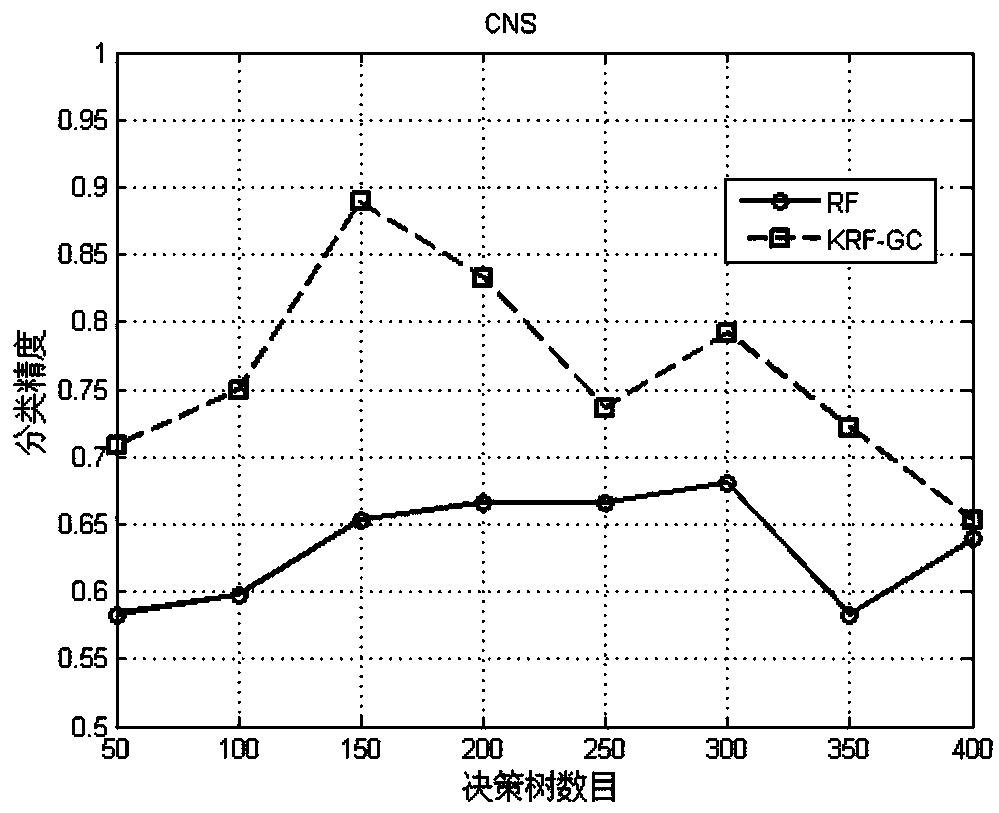
Gene classification method and system based on clustering and random forest algorithms
The invention relates to a gene classification method and system based on clustering and random forest algorithms and belongs to the technical field of biological information. The method comprises a step of acquiring gene sample data, clustering the acquired gene sample data by using the clustering algorithm to obtain a cluster center, and supplementing a training sample set with an obtained cluster center set, a step of adjusting the number of fixed decision tree random description attributes in a traditional random forest algorithm to a random value, wherein on one hand, strong decision trees in a decision tree set are kept, on the other hand, the number of average random description attributes of the decision tree set is reduced, thus the correlation between the decision trees is further reduced, and a step of predicting genetic data to be classified by using each decision tree in a random forest model. According to the method and the system, the cluster center obtained through theclustering algorithm is taken as artificial data to expand the training set of the random forest model, thus the random forest model is fully trained, the obtained classification model has high precision, and the accuracy of the classification of genetic data is improved.
Owner:HENAN NORMAL UNIV
Method for distinguishing between head and neck squamous cell carcinoma and lung squamous cell carcinoma
ActiveUS20070264644A1Sugar derivativesMicrobiological testing/measurementLung squamous cell carcinomaCXCL13
The present invention is a method distinguishing between head and neck squamous cell carcinoma and lung squamous cell carcinoma. In particular, a 10-gene classifier has been identified which can be used to distinguish between primary squamous cell carcinoma of the lung and metastatic head and neck squamous cell carcinoma. These genes include CXCL13, COL6A2, SFTPB, KRT14, TSPYL5, TMP3, KLK10, MMP1, GAS1, and MYH2. A panel of one or more of these genes, or proteins encoded thereby, can be used for early diagnosis and selection of an appropriate therapeutic treatment.
Owner:WISTAR INST THE A CORP OF PA +1
Screening method of characteristic gene of certain disease
InactiveCN101996284AReduced expression spaceReduce the effective dimensionSpecial data processing applicationsPrincipal component analysisAdditive ingredient
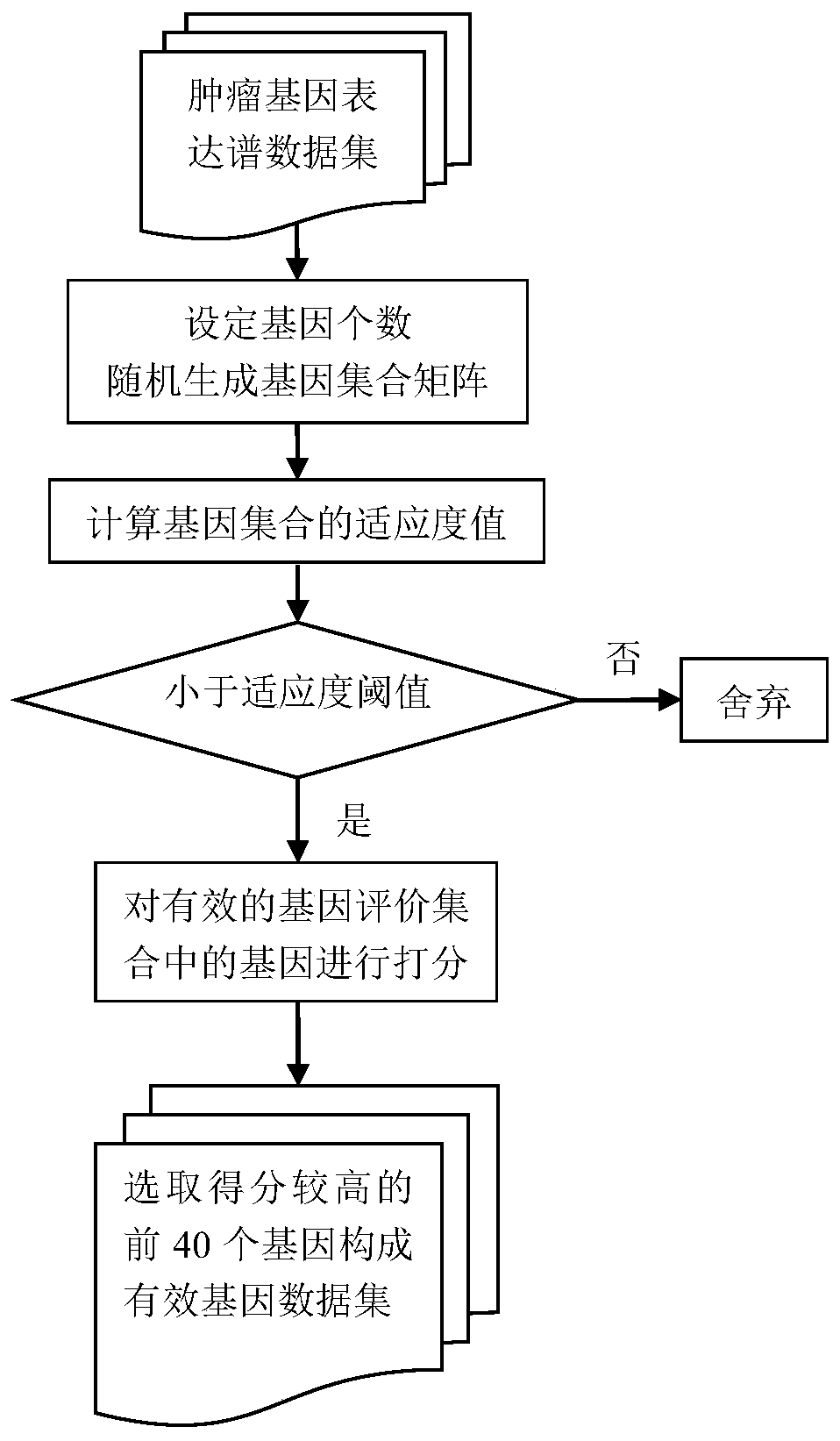
The invention provides a screening method of the characteristic gene of a certain disease. A gene expression profile is analyzed from a brand-new angle. Firstly, main ingredient analysis is carried out for reducing dimensions; under the condition that the contribution rate is 99%, a characteristic value and the contribution rate serve as classification factors to screen the characteristic gene of an oncogene and reasonably lower the effective dimensionality of gene expression space; on the basis of main ingredient analysis, Fourier transform and support vector base on the basis of a complex field effectively classify and distinguish samples; the main ingredient analysis is innovatively combined, and data processing of changing a real number field into the complex field is carried out; the frequency is recorded, and the bigger the frequency is, the better the classification result is; the gene label is reasonably and effectively extracted. The invention can be applied in the field of biological diseases, such as gene classification and distinguishing, can be applied in the field of meteorological geography, such as meteorological observation and has obvious effects and higher practical value.
Owner:KUNMING UNIV OF SCI & TECH
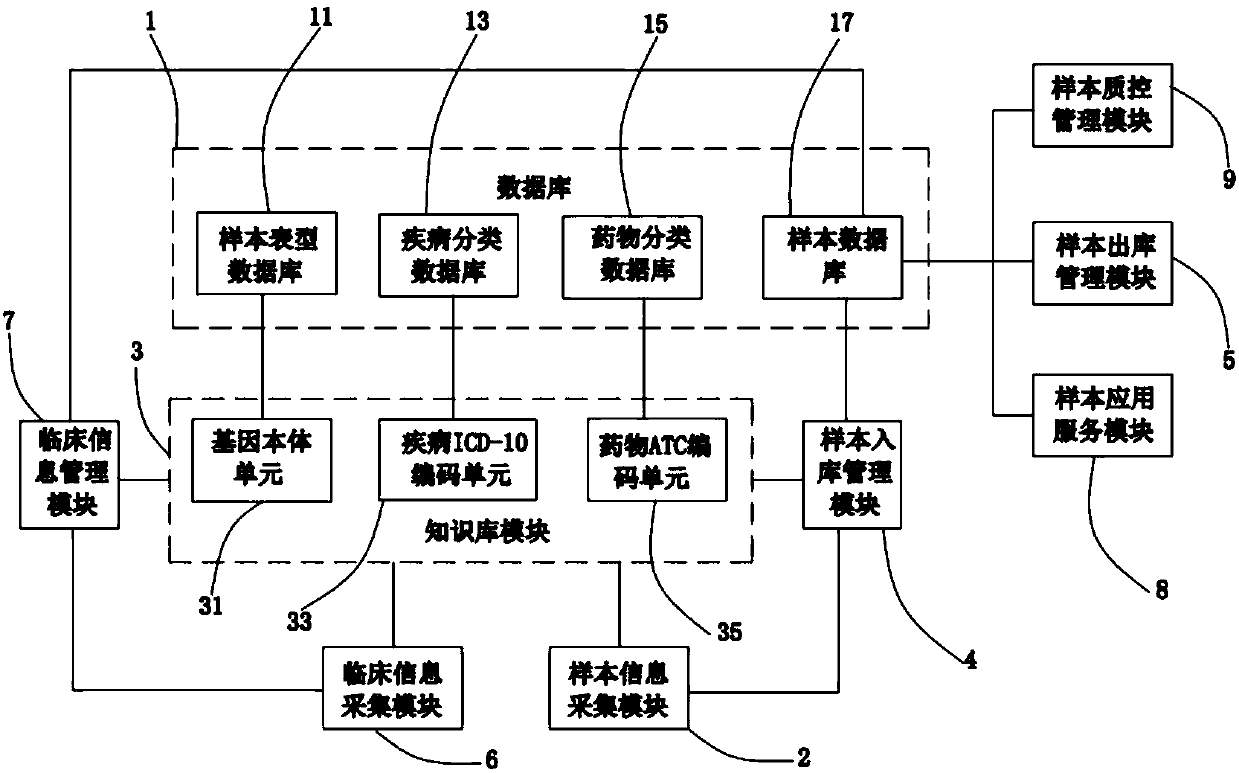
Biobank management system
InactiveCN107844685AImprove scientific research efficiencyEasy to shareProteomicsGenomicsGenomicsDisease
The invention discloses a biobank management system. The biobank management system comprises a database, a sample information acquisition module, a knowledge base module, a sample stock-in managementmodule, a sample stock-out management module, a sample application service module, a clinical sample information acquisition module and a clinical sample information management module. The biobank management system can not only achieve sample management with medicine classification or gene classification being the core, but also better meet requirements of a clinical medicine genomic clinical testduring a sample application service and can be combined with disease information to precisely stock out some sample of a patient using medicine to improve the scientific research efficiency; meanwhile, through the knowledge base module, clinical test information is standardized, not only are information sharing and transmission utilization facilitated, but also an important basis can be laid forsuccessful promoting of the clinical test and mining utilization of the information, and therefore, the clinical test efficiency can be obviously improved.
Owner:CHANGSHA 3G BIOTECH
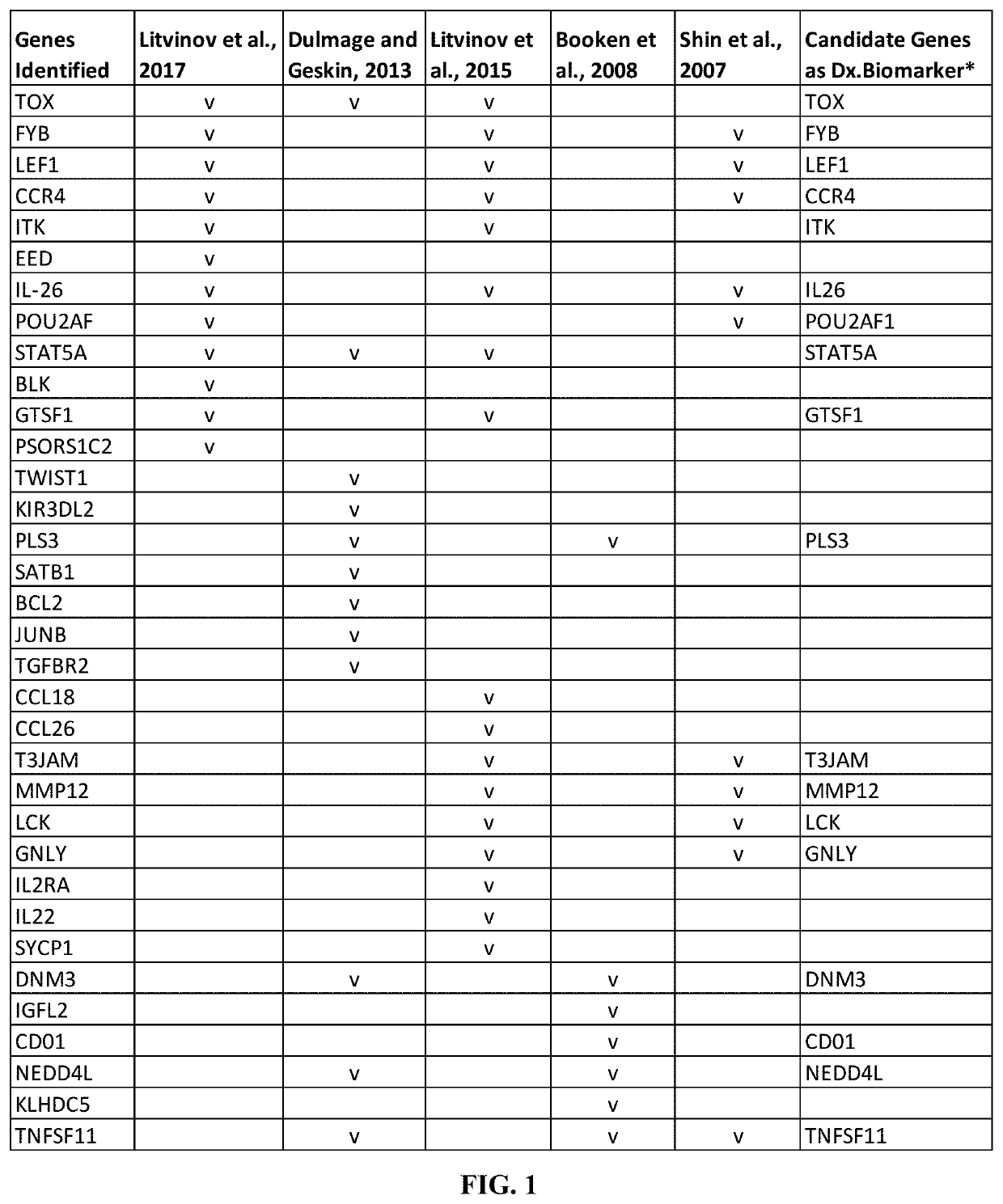
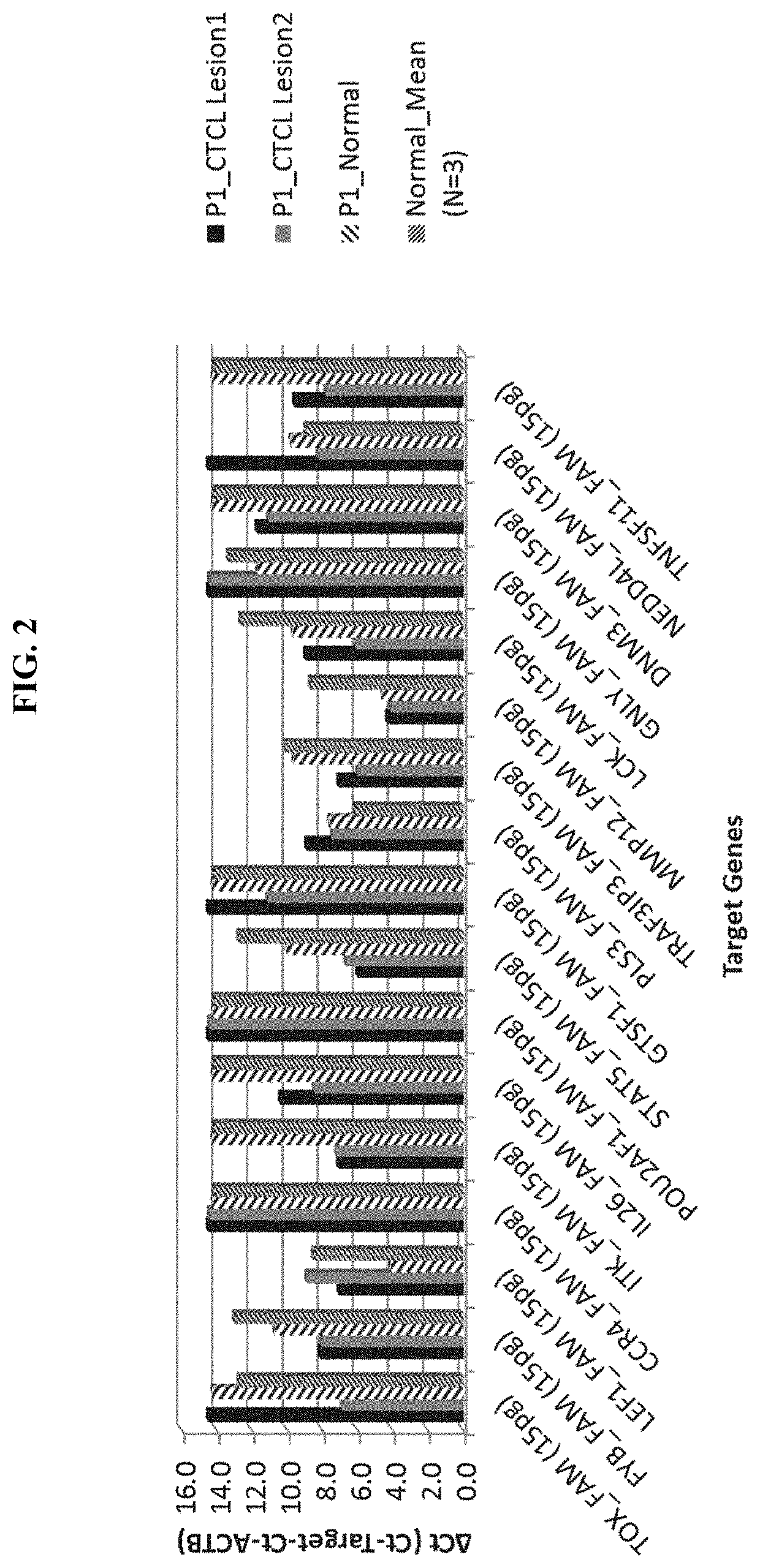
Novel gene classifiers and uses thereof in autoimmune diseases
PendingUS20200308649A1Improve treatmentMicrobiological testing/measurementSurgical needlesAutoimmune conditionMedicine
Owner:DERMTECH INC
Seedling growing method for overcoming self-incompatibility of pears
InactiveCN103250573AAvoid major economic lossesCultivating equipmentsHorticultureRootstockPollination
Provided is a seedling growing method for overcoming self-incompatibility of pears. The method includes the steps of stock seedling breeding and grafting. According to the step of grafting, two varieties are selected and combined as cions, and the two varieties are different in S gene classification, synchronous in flowering phase, large in pollen quantity and strong in pollen viability; in breeding of pear variety seedlings without pollen, the pear variety seedlings need to be combined with other two varieties or three varieties as cions, and the two varieties or three varieties are different in S gene classification, synchronous in flowering phase, large in pollen quantity and strong in pollen viability; the grafting method is bud grafting, the combinations of the two varieties are Yuan Huang (S3S4)+Cui Guan (S2S5), and the like, and the combinations of the three varieties is Xin Gao (S3S9)+Yuan Huang (S3S4)+Cui Guan (S2S5), and the like. According to the seedling growing method for overcoming the self-incompatibility of the pears, problems existing in pollination variety selective breeding of the pears are fundamental solved in the seedling breeding stage, and therefore great economic losses brought to orchard workers in production and caused by the self-incompatibility of the pears can be avoided.
Owner:INST OF FRUIT & TEA HUBEI ACAD OF AGRI SCI
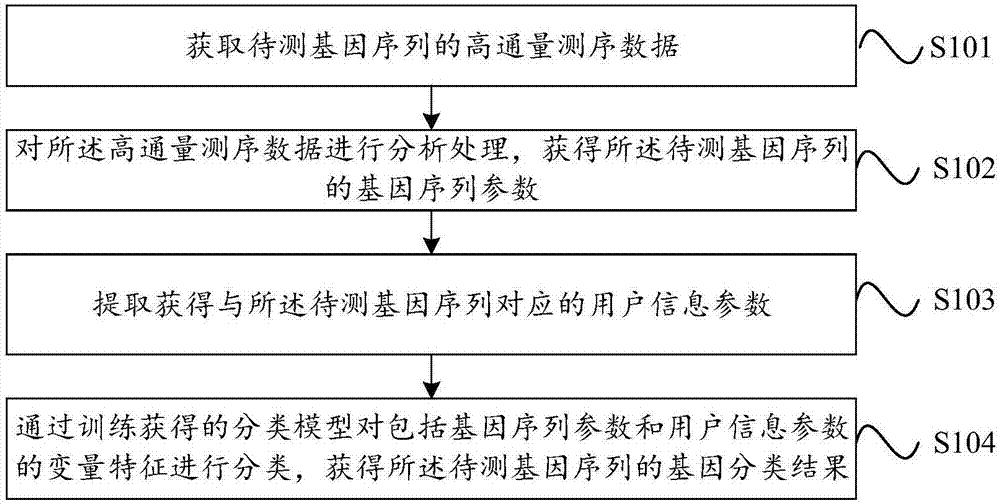
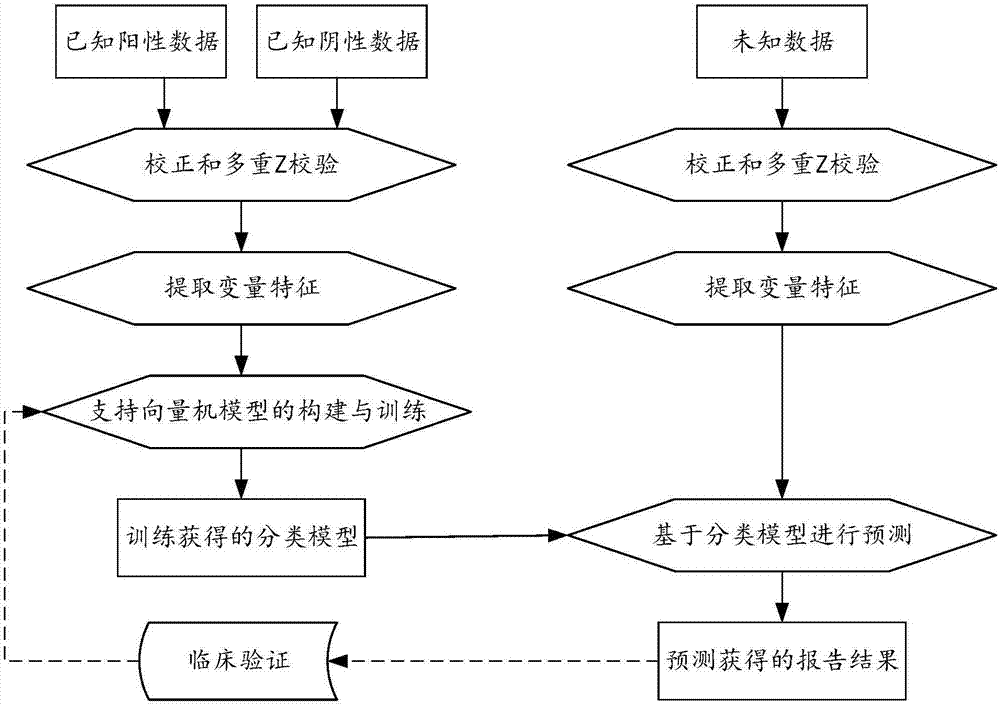
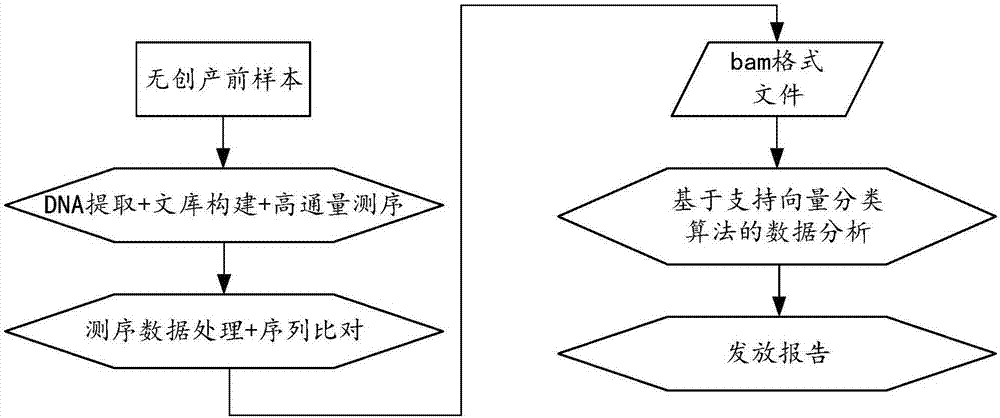
Biological information analysis method and device for high-throughput sequencing, equipment and storage medium
ActiveCN107463797AImprove accuracyReduce testing costsHybridisationSpecial data processing applicationsInformation analysisVariable features
The invention discloses a biological information analysis method and device for high-throughput sequencing, equipment and a storage medium. The method in one embodiment comprises the following steps of: obtaining high-throughput sequencing data of a to-be-tested gene sequence; analyzing the high-throughput sequencing data to obtain a gene sequence parameter of the to-be-tested gene sequence; extracting a user information parameter corresponding to the to-be-tested gene sequence; and classifying variable features comprising the gene sequence parameter and the user information parameter through a classification model obtained through training so as to obtain a gene classification result of the to-be-tested gene sequence. According to the method, the detection cost is reduced, and false positive and negative results occurring before can be corrected at the same time, so that the detection correctness is improved.
Owner:广州达安临床检验中心有限公司 +4
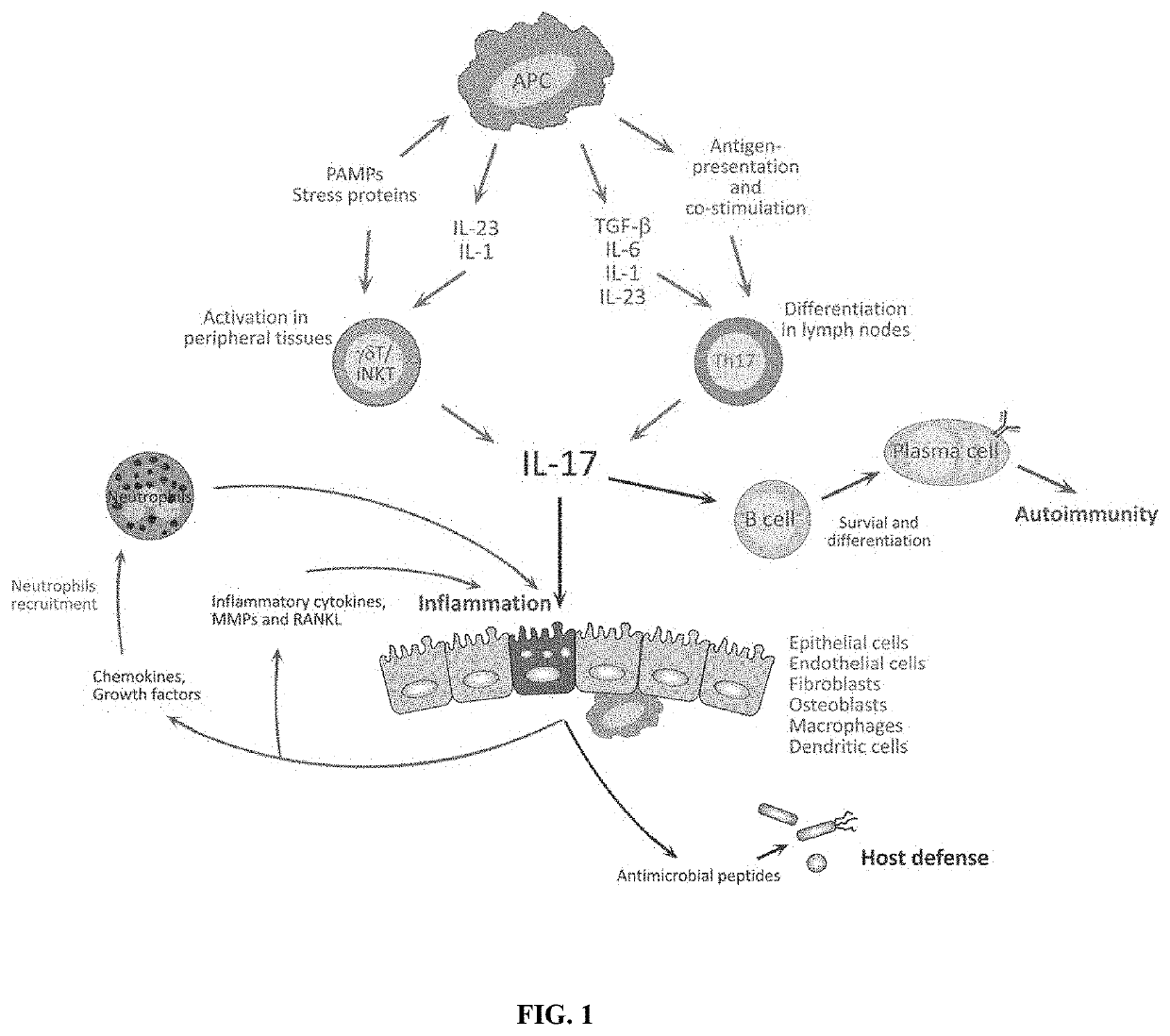
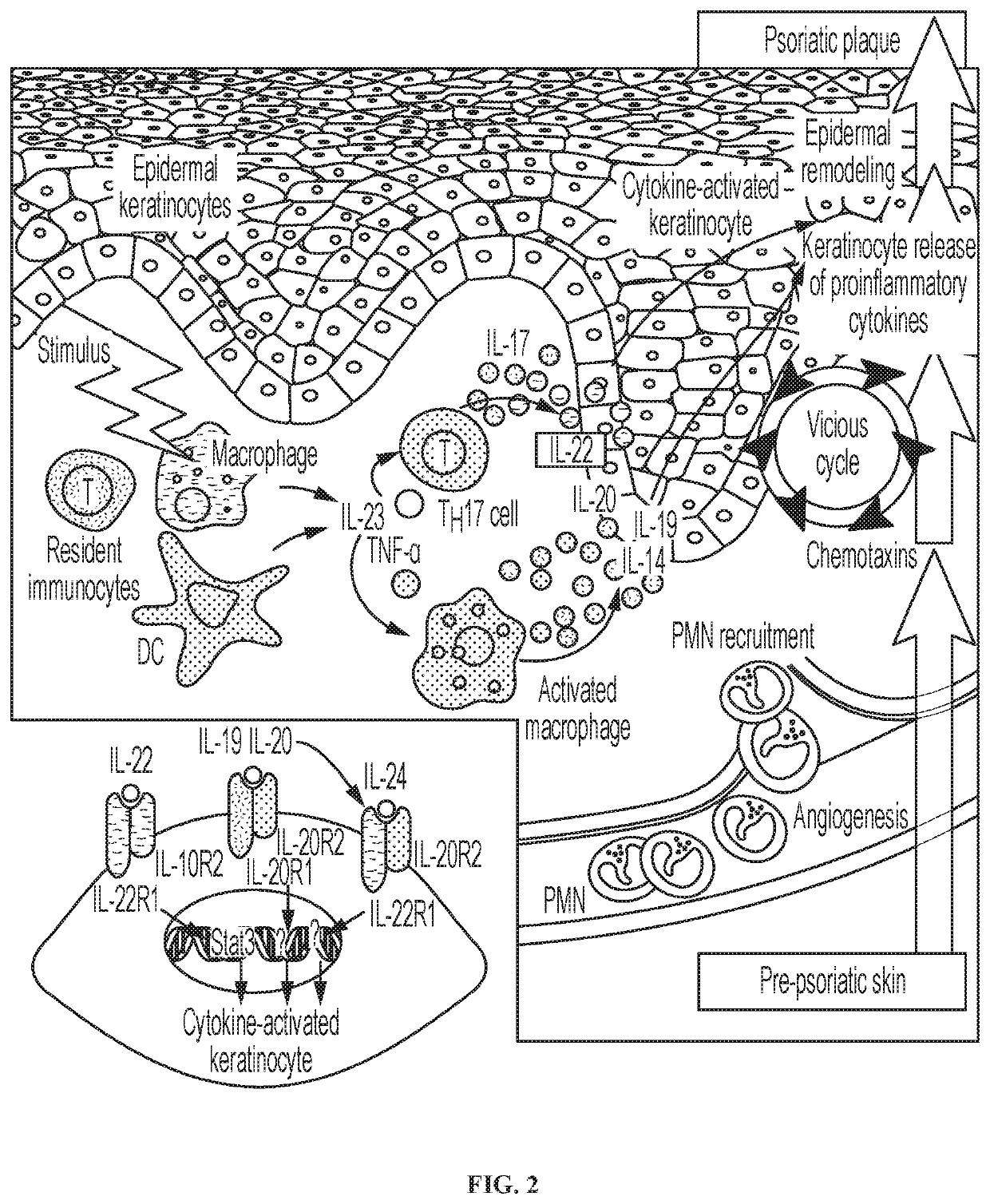
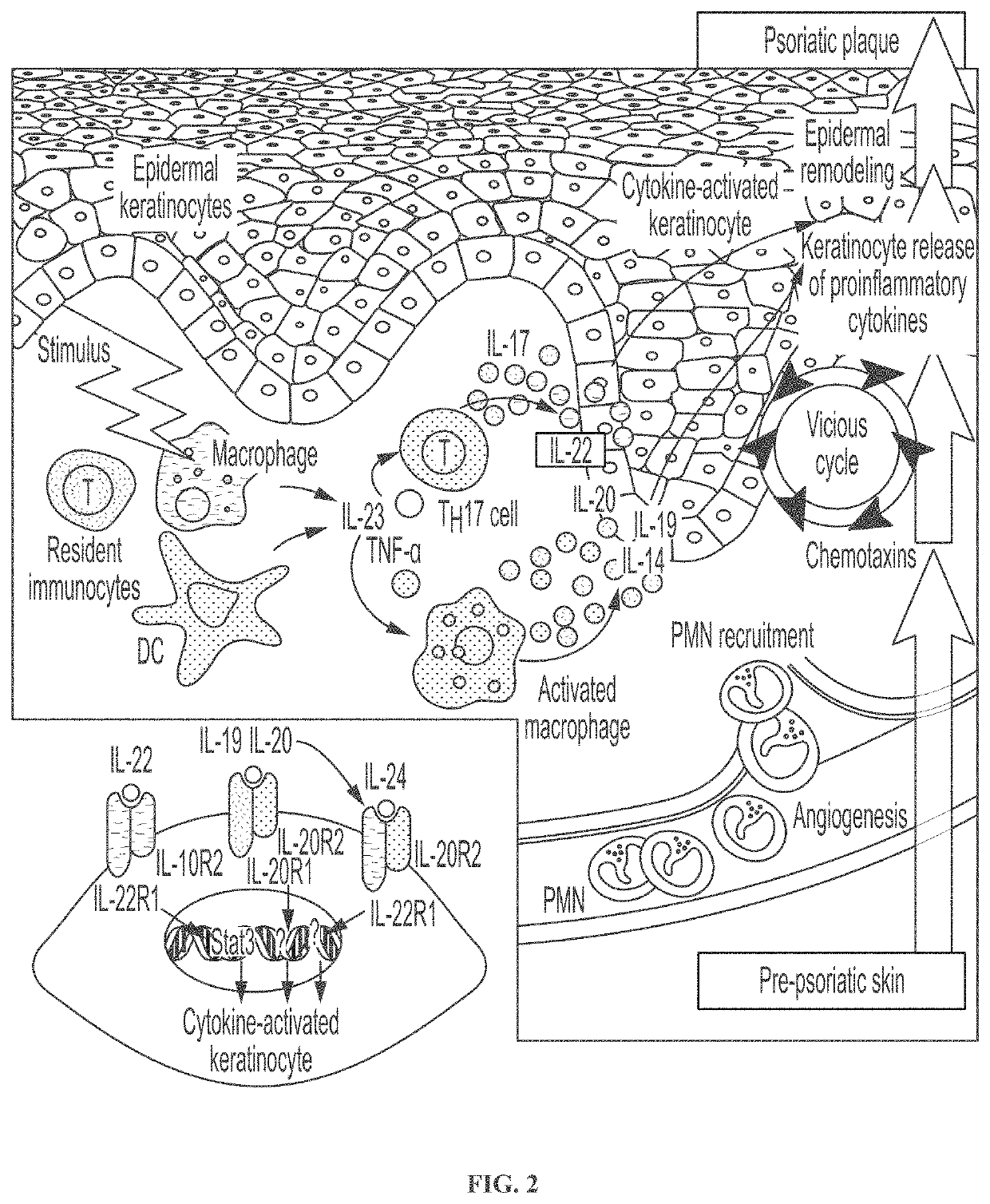
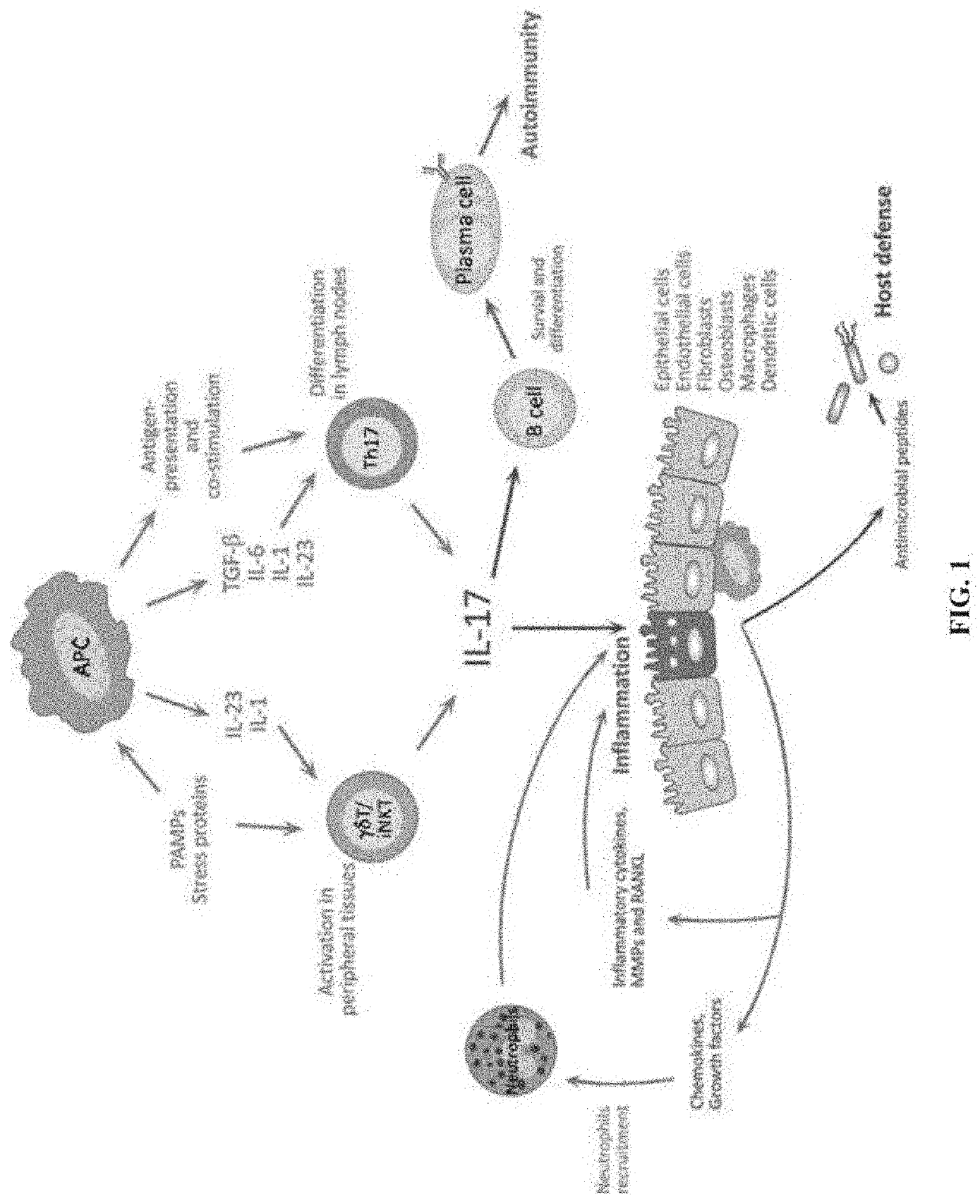
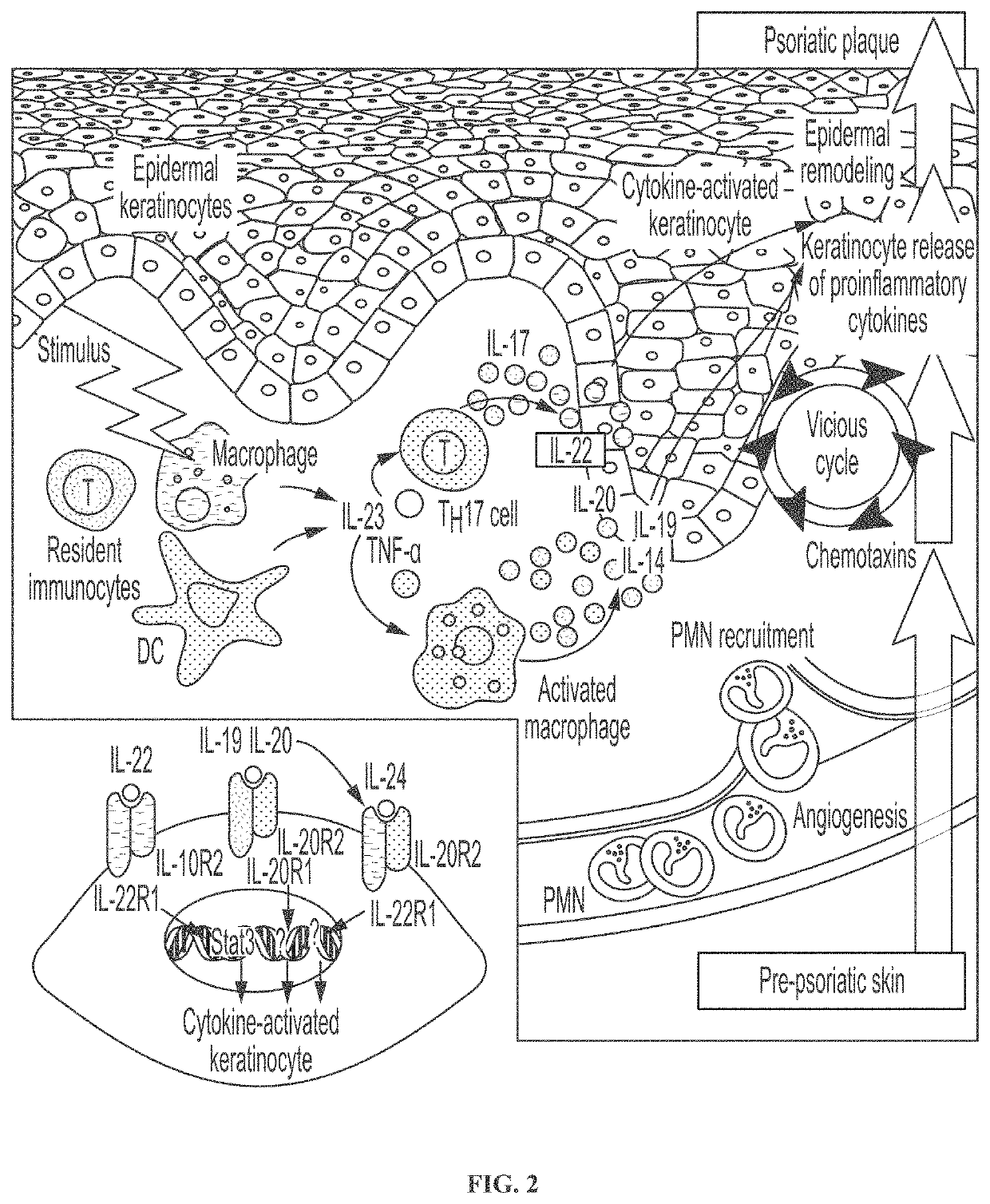
Novel gene classifiers and uses thereof in psoriasis
PendingUS20210222247A1Microbiological testing/measurementSurgical needlesMedicineGene expression level
Owner:DERMTECH INC
Novel gene classifiers and uses thereof in skin cancers
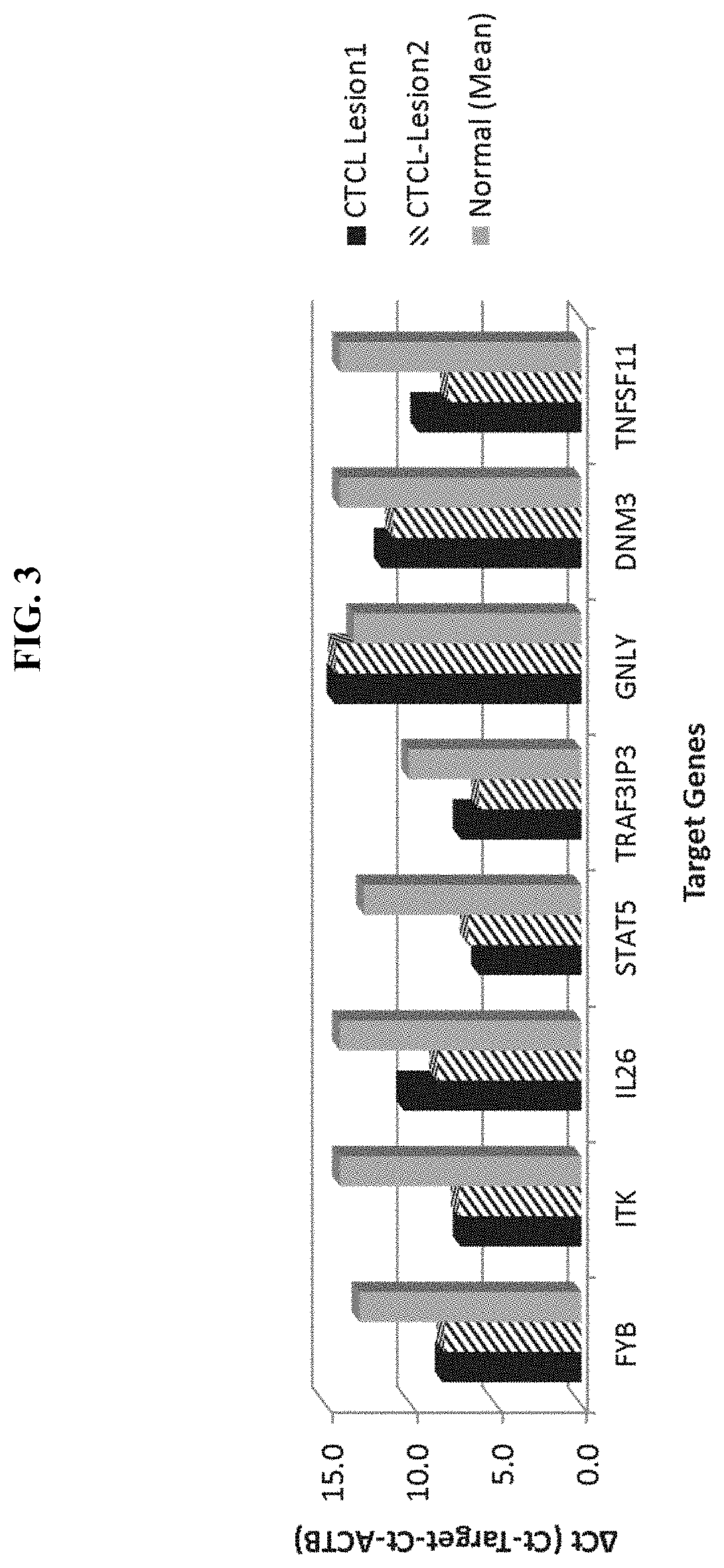
ActiveUS20200308657A1Organic active ingredientsPeptide/protein ingredientsGranulomaCutaneous T-cell lymphoma
Owner:DERMTECH INC
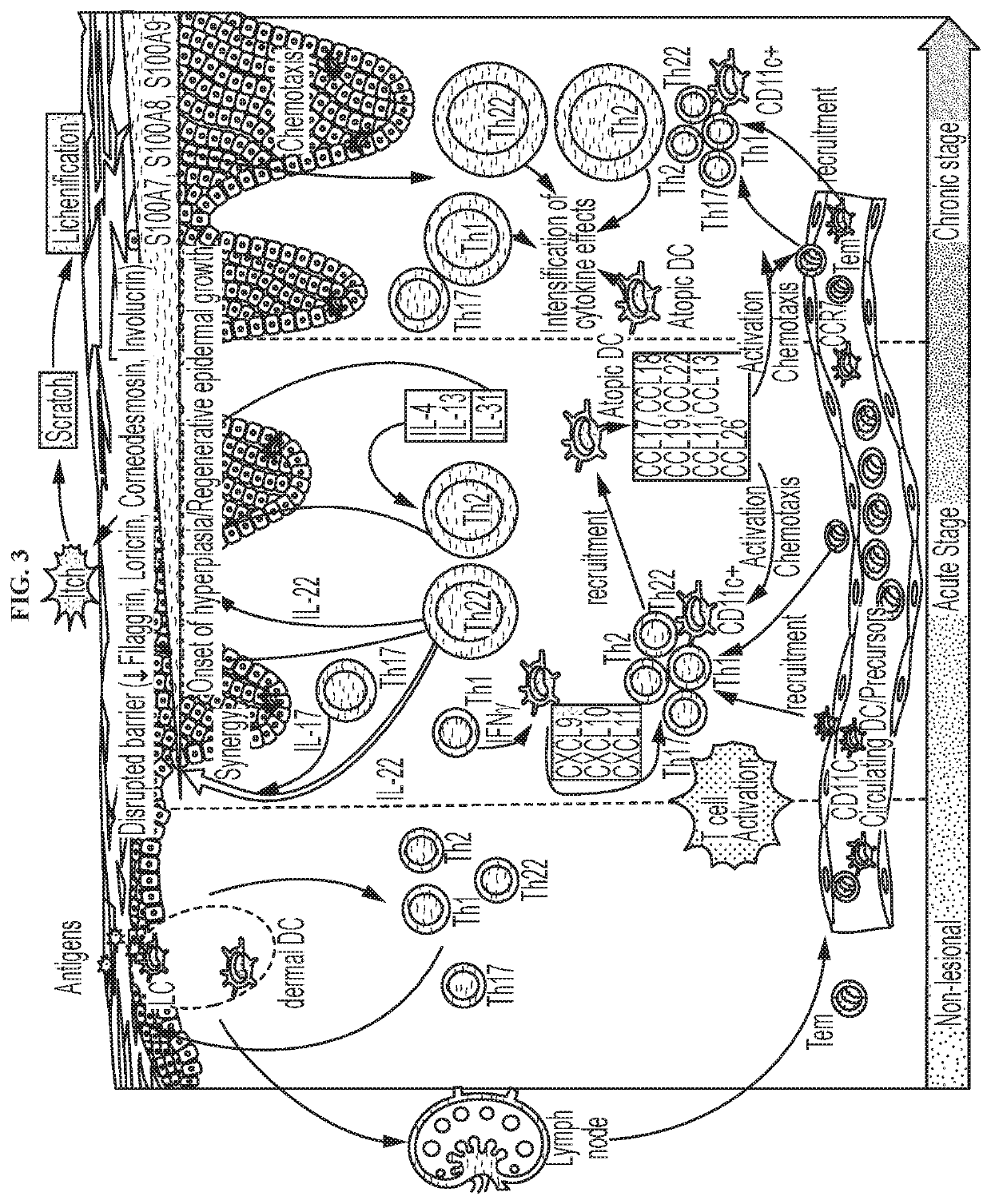
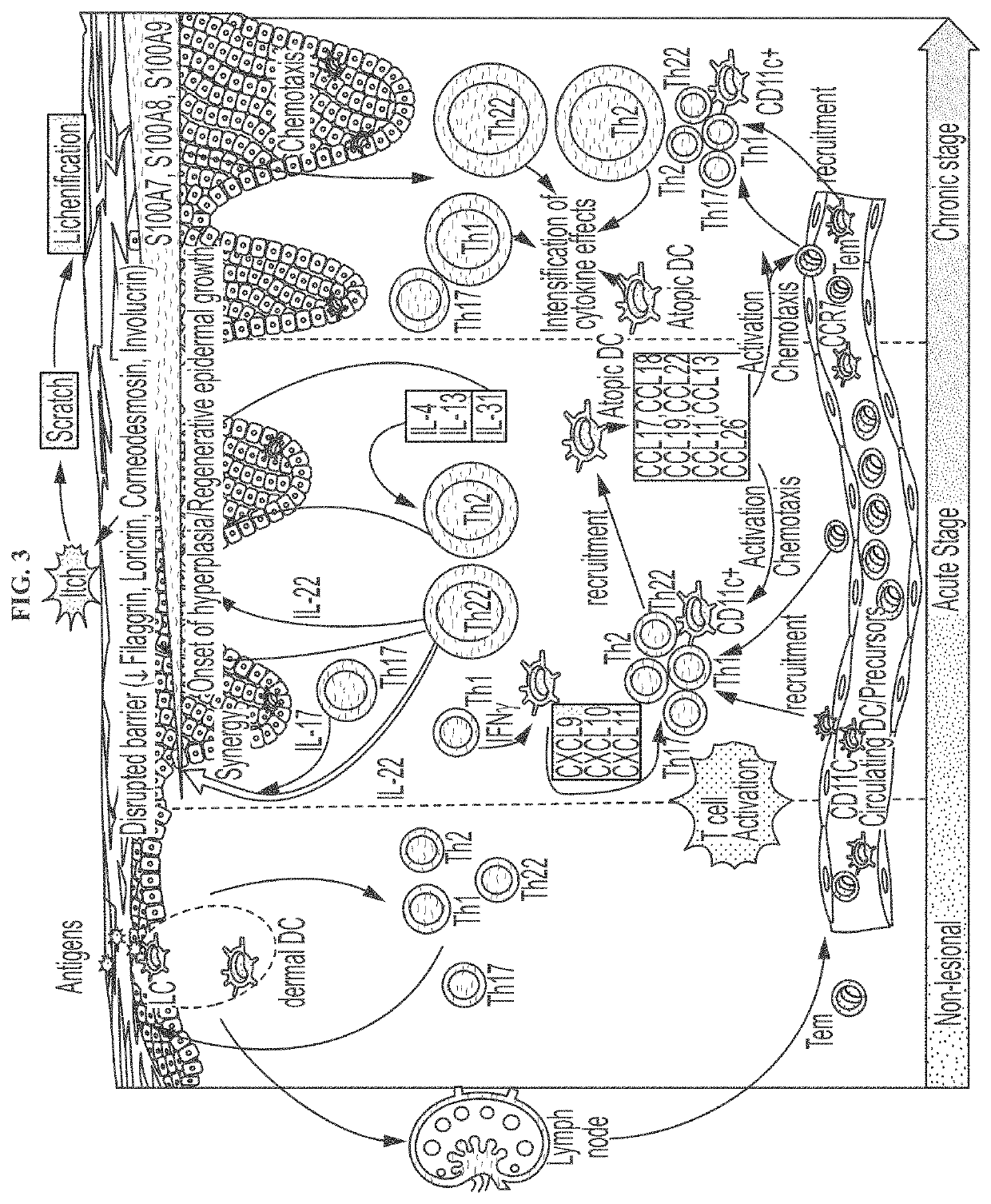
Novel gene classifiers and uses thereof in atopic dermatitis
ActiveUS20210222246A1Microbiological testing/measurementSurgical needlesAtopic dermatitisExpression gene
Disclosed herein are methods of detecting an altered gene expression levels in a subject suspected of having atopic dermatitis. Further described herein are methods of treating atopic dermatitis in a subject having an exhibiting an altered gene expression level.
Owner:DERMTECH INC
Gene classifiers for use in monitoring UV damage
PendingUS20200319205A1Level of expressionMicrobiological testing/measurementDisease diagnosisMedicineComputer science
Disclosed herein is a method of detecting the presence of skin UV damage based on molecular risk factors. In some instances, also described herein is a method of determining the progression of UV damage based on the molecular risk factors.
Owner:DERMTECH INC
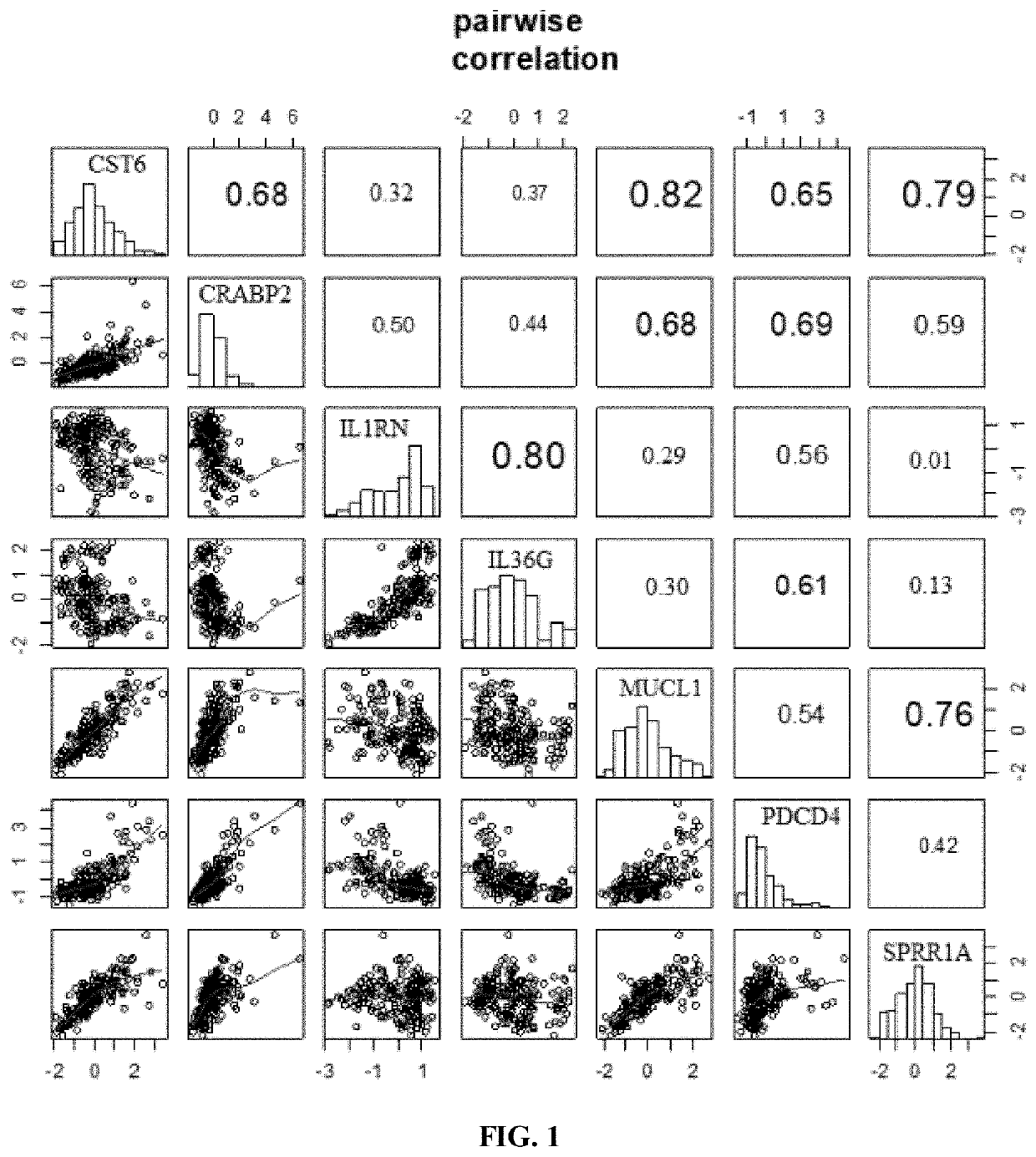
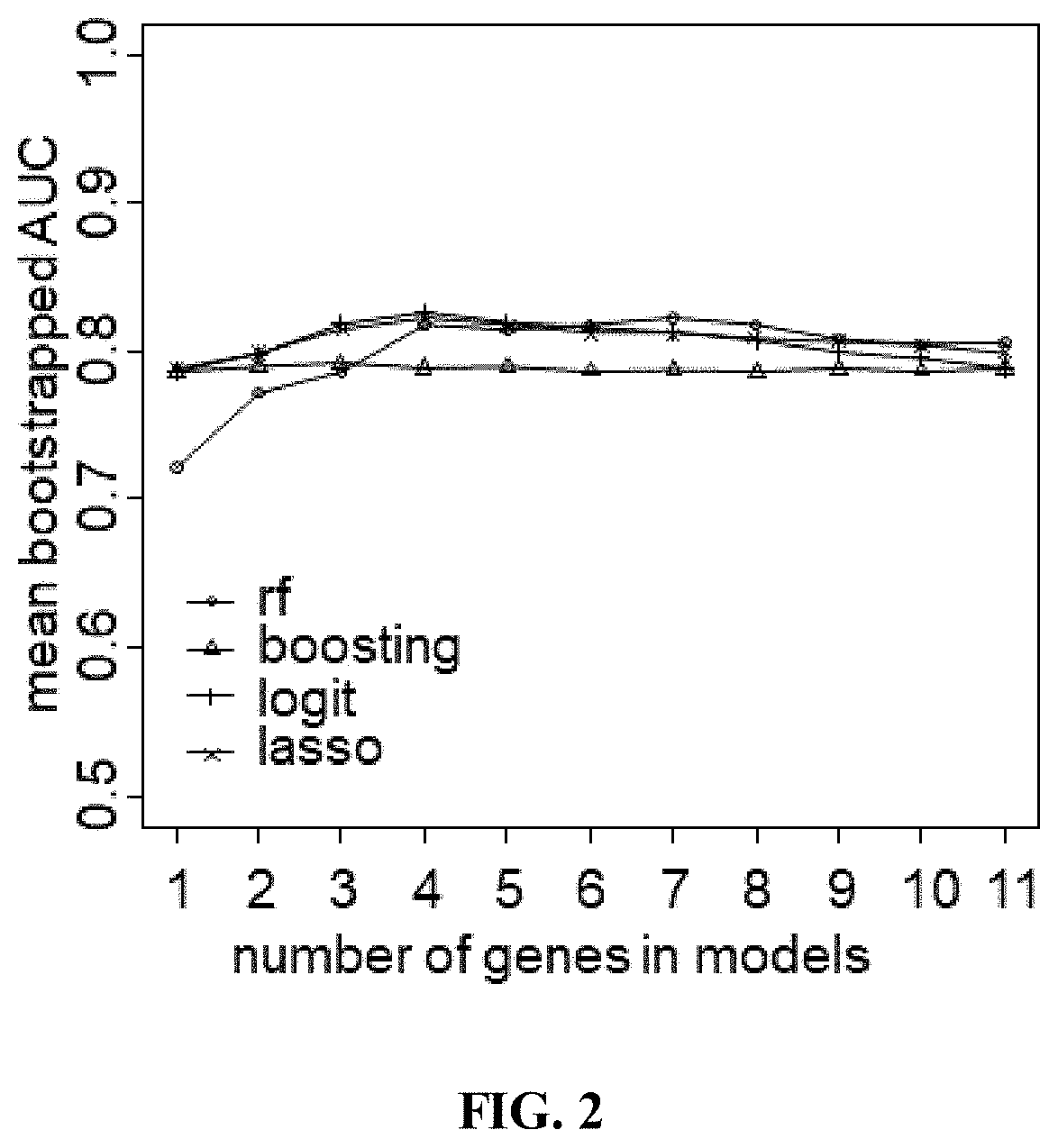
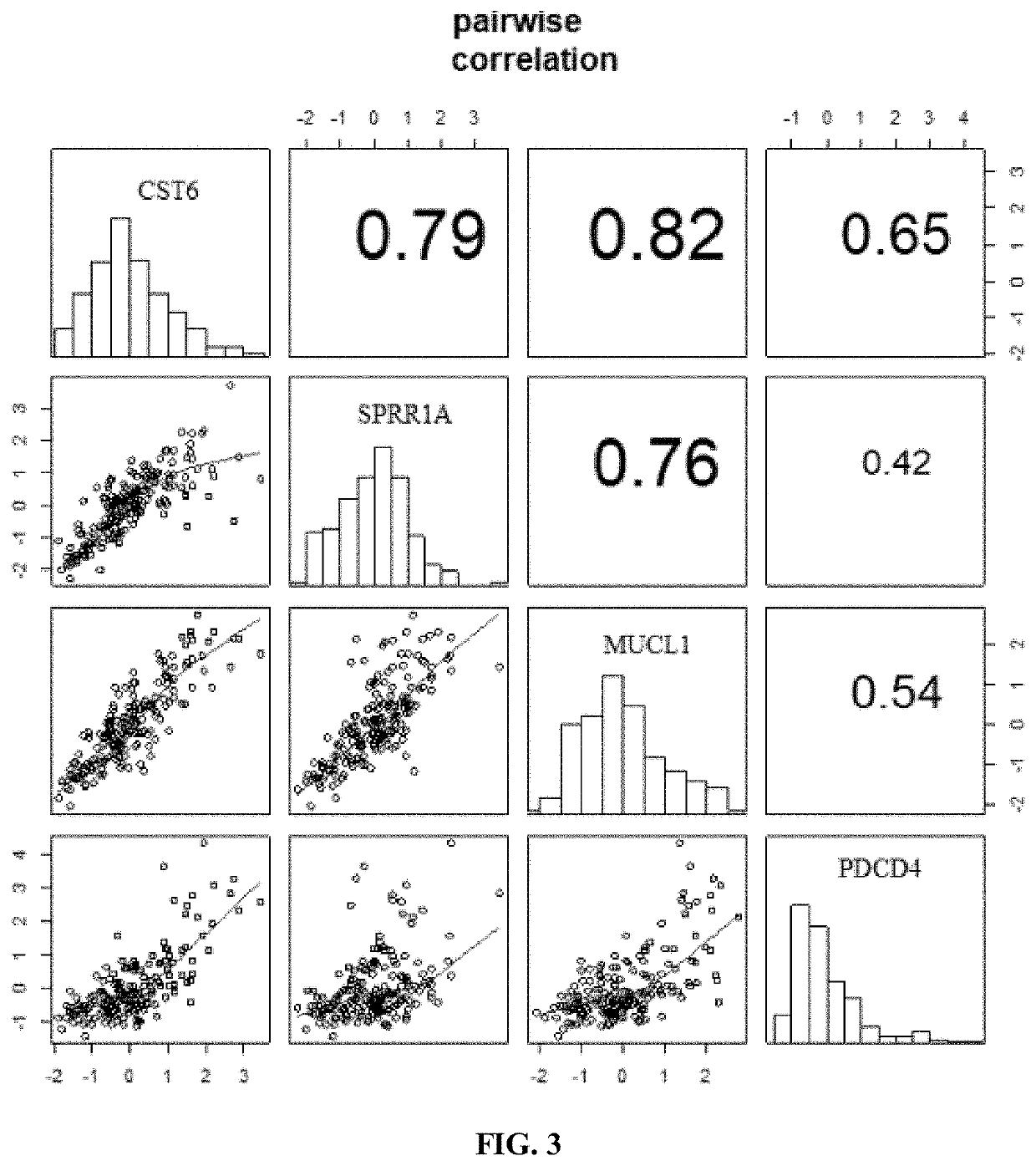
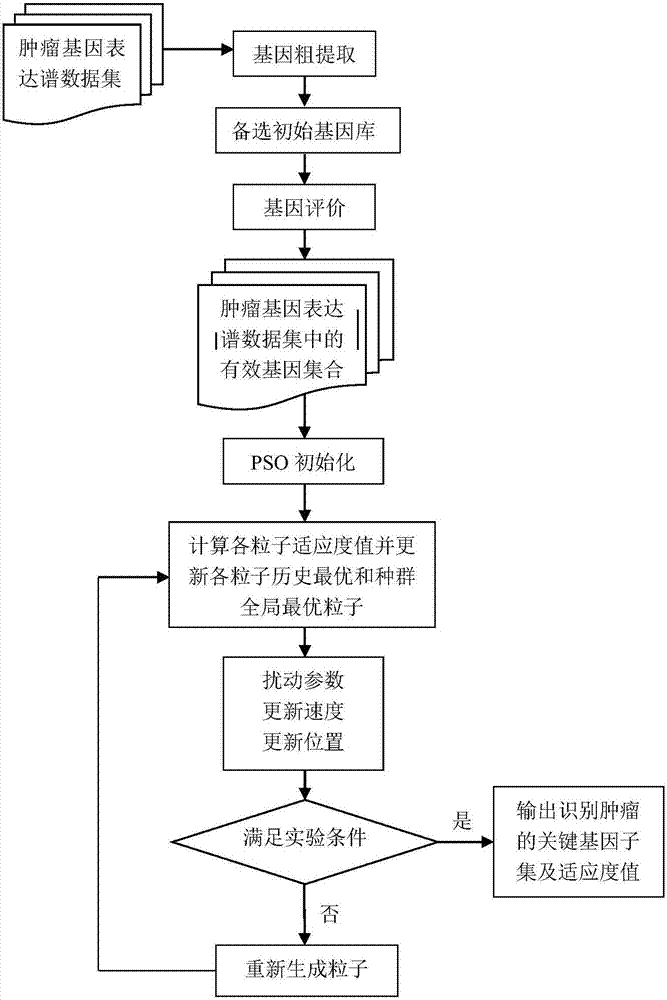
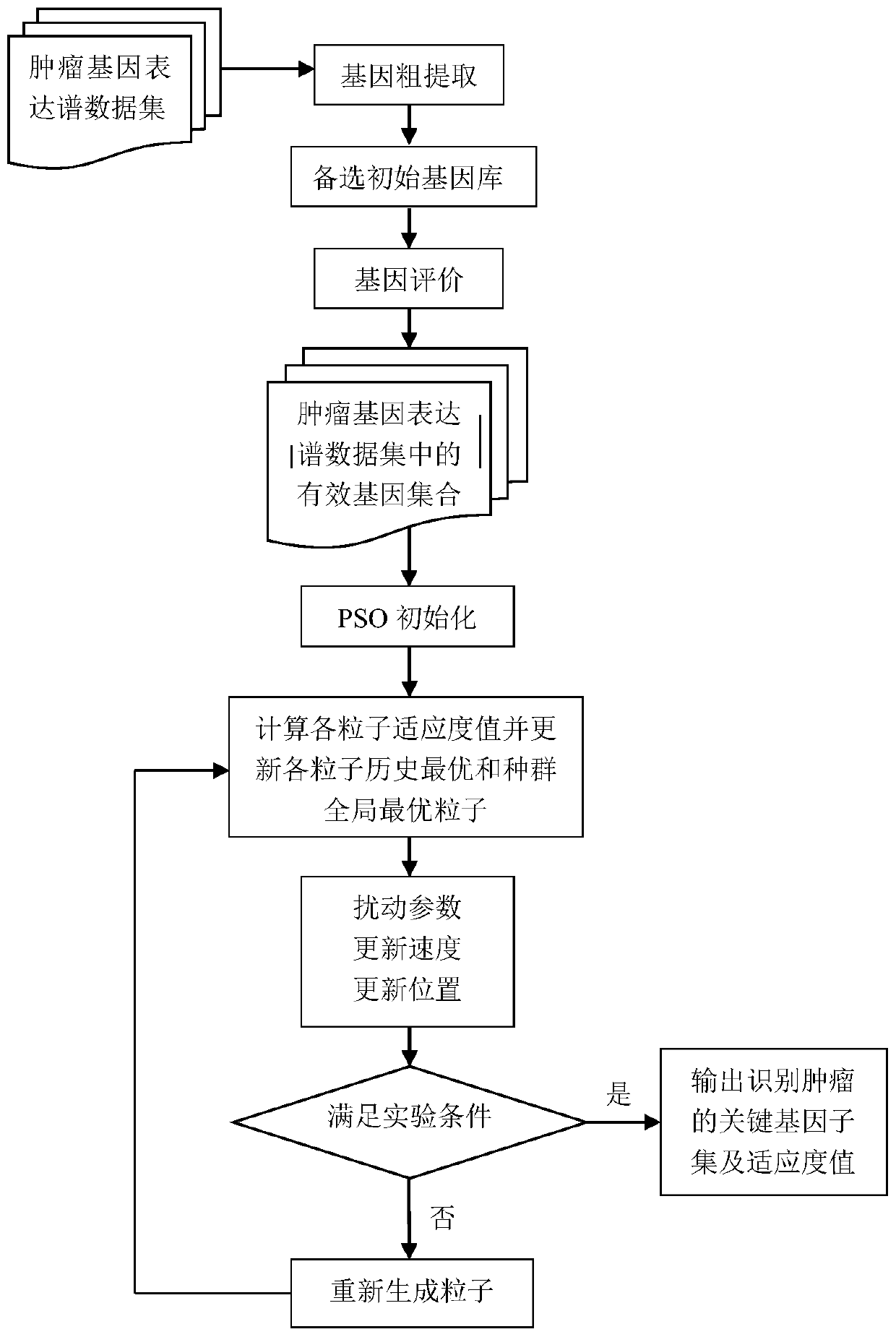
Tumor key gene identification method based on particle swarm optimization and marking criterion
ActiveCN106951728AAccurate identificationEffectively deleteProteomicsGenomicsLocal optimumMulti-swarm optimization
The invention discloses a tumor key gene identification method based on particle swarm optimization and a marking criterion. The method comprises the steps that one gene marking criterion is defined to acquire gene classification ability information so as to filter genes with a low correlation to a tumor category; and the Metropolis criterion is utilized to improve a particle swarm optimization (PSO) algorithm in combination with the gene classification ability information so as to realize identification of tumor key genes. The new method for gene classification information and PSO improvement overcomes the defect that a traditional method for identifying tumor key genes based on PSO is prone to fall into a locally optimal solution, gene subsets in a smaller number and with a high correlation to the tumor category can be selected, and therefore the method is beneficial for improving subsequent tumor identification.
Owner:JIANGSU UNIV
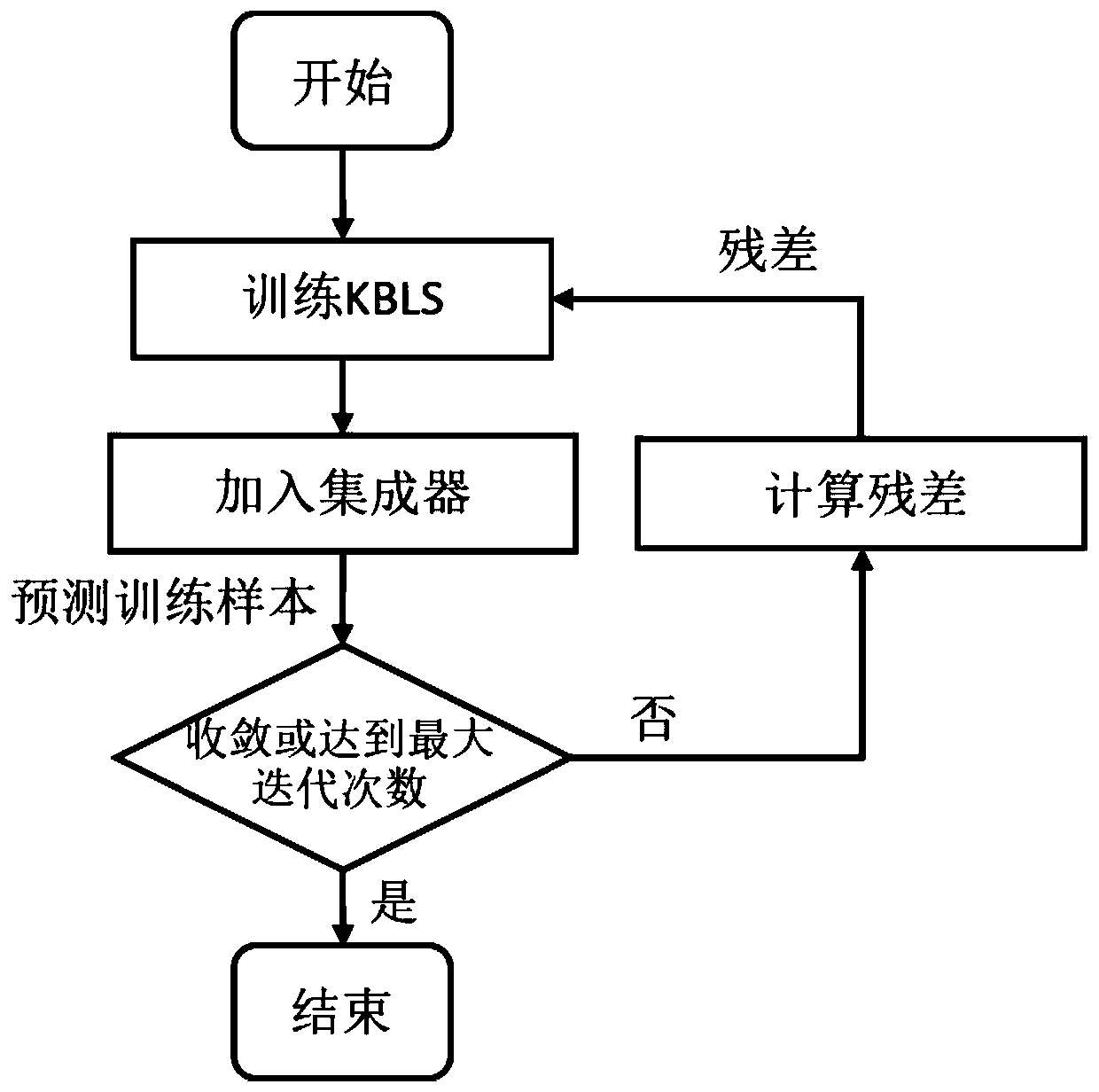
Progressive ensemble classification method based on kernel width learning system
PendingCN111598187ANoise resistance characteristicsImprove classification performanceEnsemble learningCharacter and pattern recognitionData setTest sample
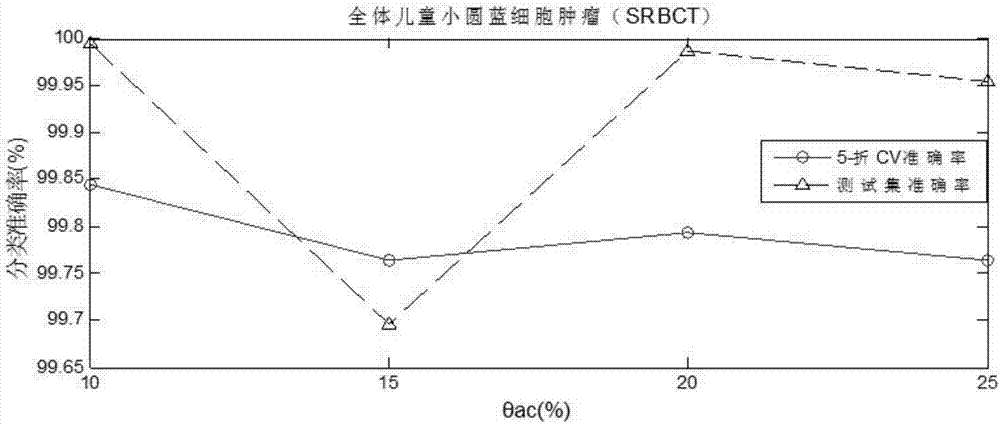
The invention discloses a progressive ensemble classification method based on a kernel width learning system. The progressive ensemble classification method comprises the following steps: 1) inputtinga training sample and a test sample; 2) training a kernel width learning system as a base classifier by using the original training data; 3) calculating a prediction residual error according to the training result of the first base classifier, and taking the prediction residual error as a label trained by the next base classifier; 4) when the reduction rate of the trained loss function value reaches a threshold value, stopping the training, and not continuing to increase the base classifier any more; and 5) classifying the test samples to obtain a final prediction result. According to the method, the nonlinear fitting capability of the classifier is improved by introducing a kernel mapping technology while redundant back propagation is not needed by utilizing a width learning system, anda plurality of base classifiers are fused by using an integrated means, so that an obvious improvement effect is achieved on a biological information data set with noise, and the accuracy of biological gene classification is improved.
Owner:SOUTH CHINA UNIV OF TECH
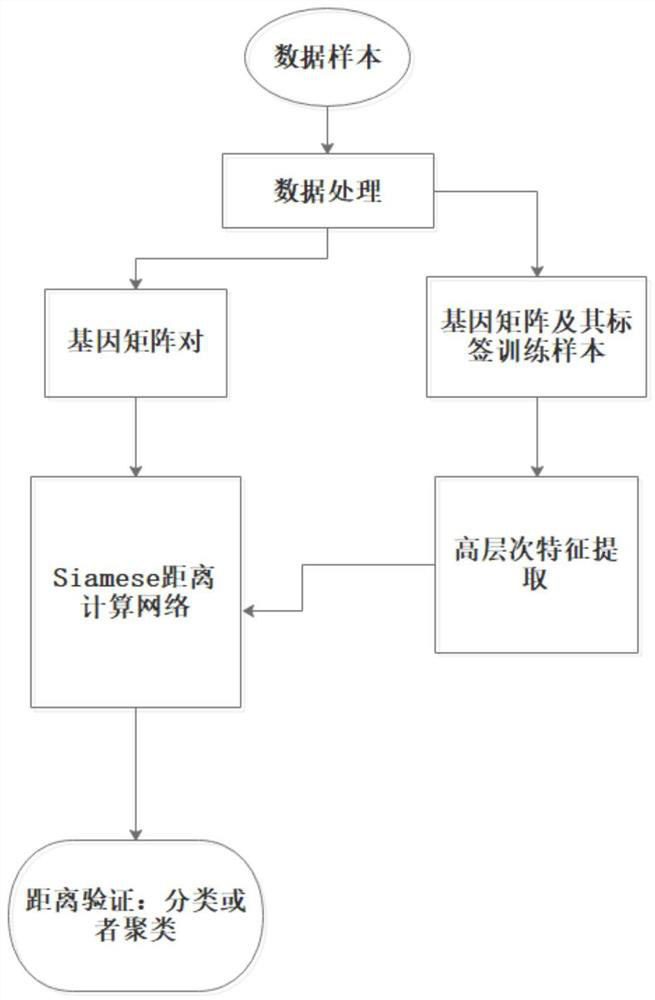
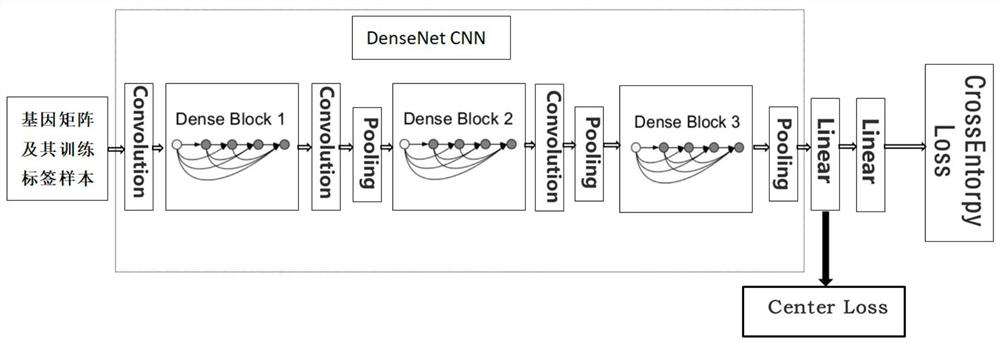
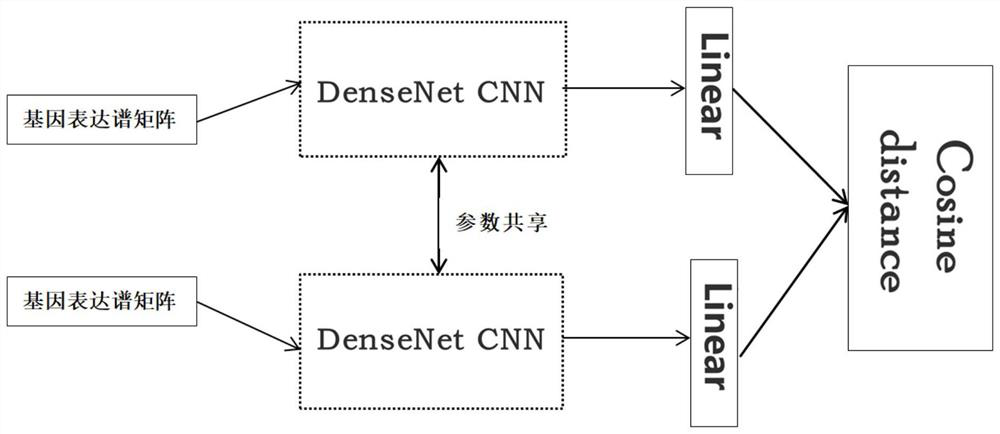
Gene expression profile distance measurement method based on deep learning
ActiveCN110033041AAccurate and fast calculationCharacter and pattern recognitionNeural architecturesGene EnrichmentProfile analysis
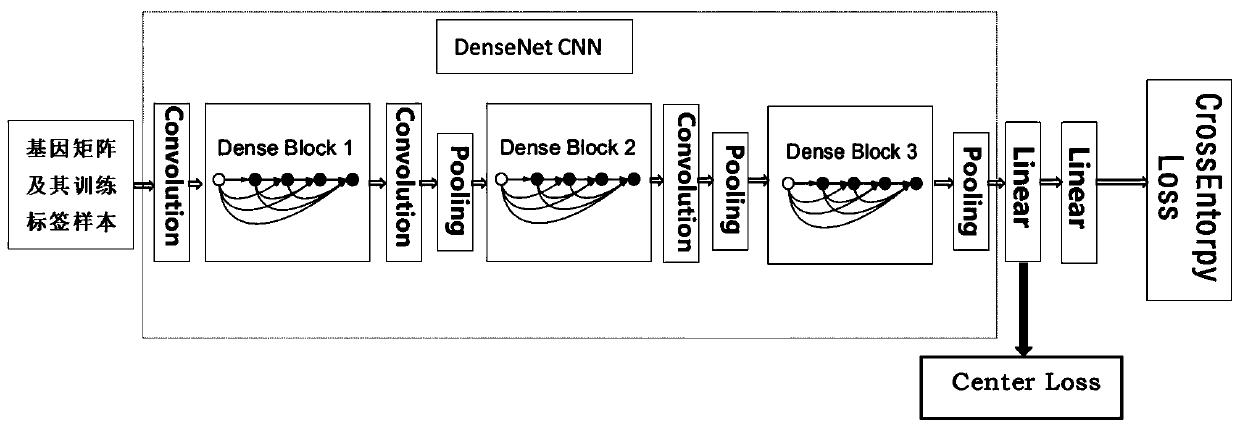
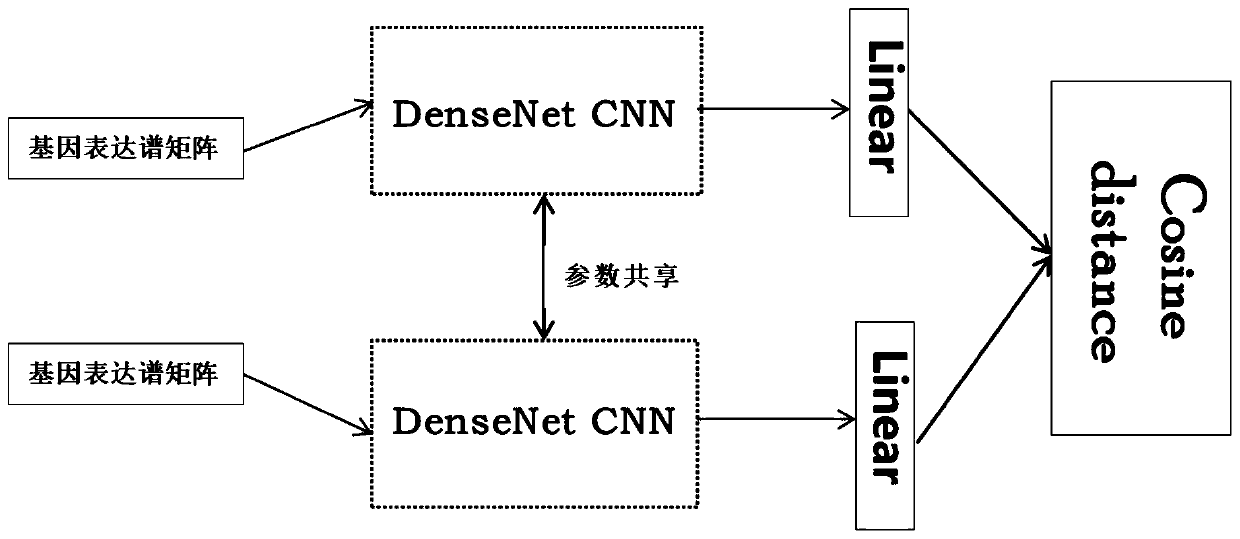
The invention belongs to the field of gene expression profile classification, discloses a gene expression profile distance measurement method based on deep learning, and belongs to mining and application of deep learning on biological big data. Firstly, a convolutional neural network model suitable for gene characteristic metric learning is designed to extract data characteristics, then the distance between the data is calculated by applying an improved cosine distance, and finally, the good performance of the method is measured through the classification effect of a classification algorithm.According to the method, the similarity between different gene expression profiles can be quickly and efficiently measured, and data is provided for subsequent researches such as gene classification,clustering, differential expression analysis and compound screening. Compared with a traditional gene enrichment method, the method has the advantages that the distance measurement effect between thedata is obviously improved, the manual intervention during gene expression profile analysis can be effectively reduced, the overfitting phenomenon easily generated by a conventional deep network is avoided, and the method has relatively high mobility.
Owner:HUNAN UNIV
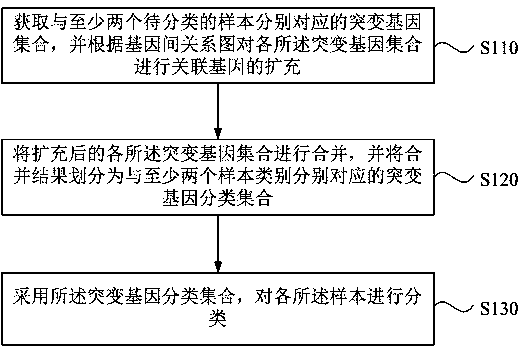
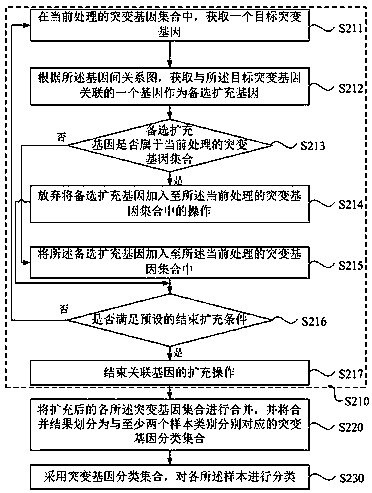
Method, apparatus, device and medium for sample classification based on gene mutation information
InactiveCN109411018AAddressing the lack of biological significanceSolve the problem of poor classification effectBiostatisticsProteomicsCurrent sampleBioinformatics
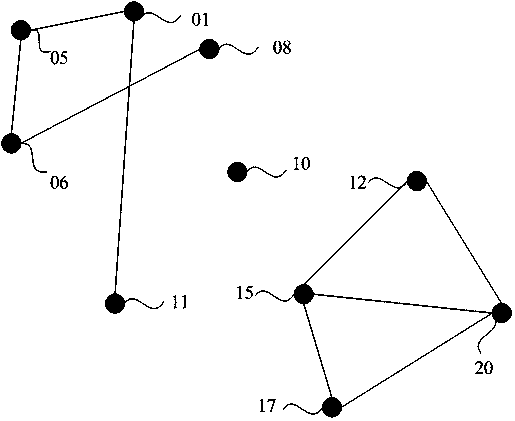
The invention discloses a method, an apparatus, a device and a medium for sample classification based on gene mutation information. The method comprises the steps that mutated gene sets correspondingto at least two samples to be classified are obtained, and expansion of associated gene is conducted on the mutated gene sets based on a gene relation pattern; the expanded mutated gene sets are merged, and the merging result is divided into mutated gene classification sets, respectively corresponding to at least two sample types; the mutated gene classification set is adopted to conduct classification on the sample. The invention is advantageous in that the current sample classification mode based on mutated gene can be optimized, and thereby classification accuracy can be improved.
Owner:SHANGHAI BIOTECAN PHARMA +1
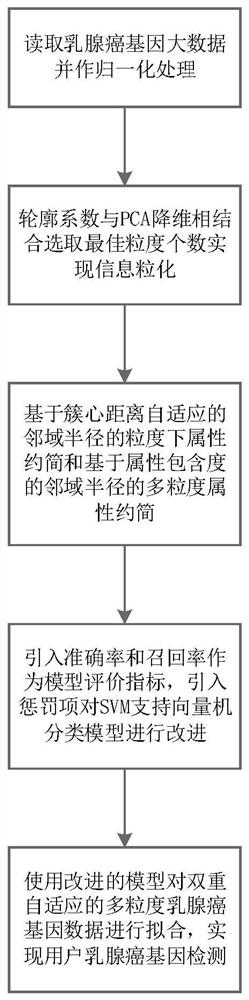
Multi-granularity breast cancer gene classification method based on dual adaptive neighborhood radius
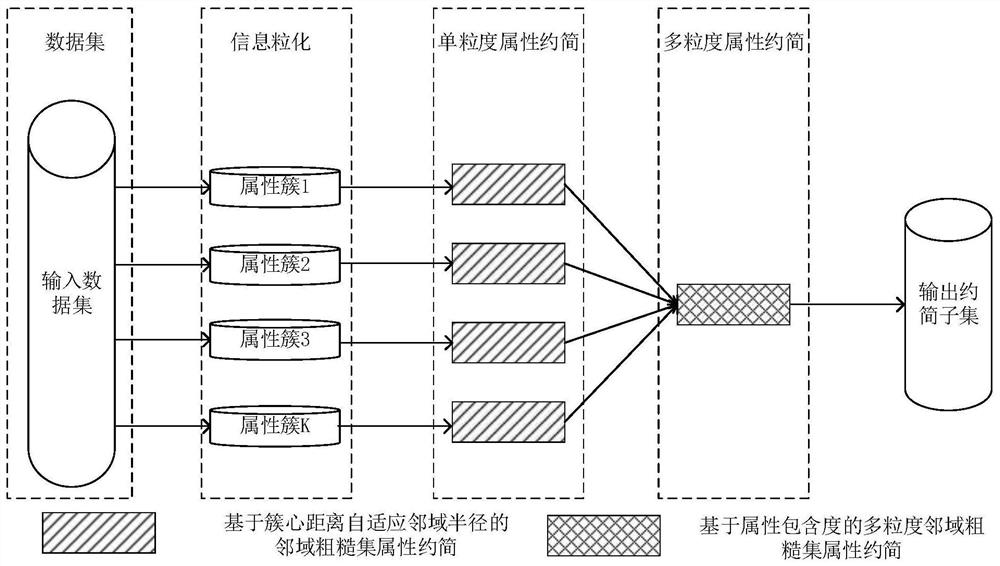
ActiveCN113838532AImprove accuracyBest treatment periodMedical data miningBiostatisticsSupport vector machineLarge scale data
The invention provides a multi-granularity breast cancer gene classification method based on dual adaptive neighborhood radiuses, and the method comprises the following steps: reading large-scale gene locus data, carrying out normalization processing, and carrying out data analysis on large-scale gene loci; combining a contour coefficient and PCA dimension reduction visualization, selecting an optimal K value, and adjusting an information granulation model; realizing the multi-granularity attribute reduction based on cluster center distance adaptive neighborhood radius and multi-granularity attribute reduction based on neighborhood radius of attribute inclusion degree by using a heuristic reduction algorithm, and classifying and predicting breast cancer gene big data are classified and predicted by adopting an SVM machine learning classification algorithm. The invention has the beneficial effects that the penalty term is adjusted, so that the model has higher accuracy and recall rate in breast cancer gene classification, redundant attributes in large-scale data are removed, the calculation efficiency is improved, and the efficiency and precision of breast cancer data classification are improved by utilizing support information between samples.
Owner:NANTONG UNIVERSITY
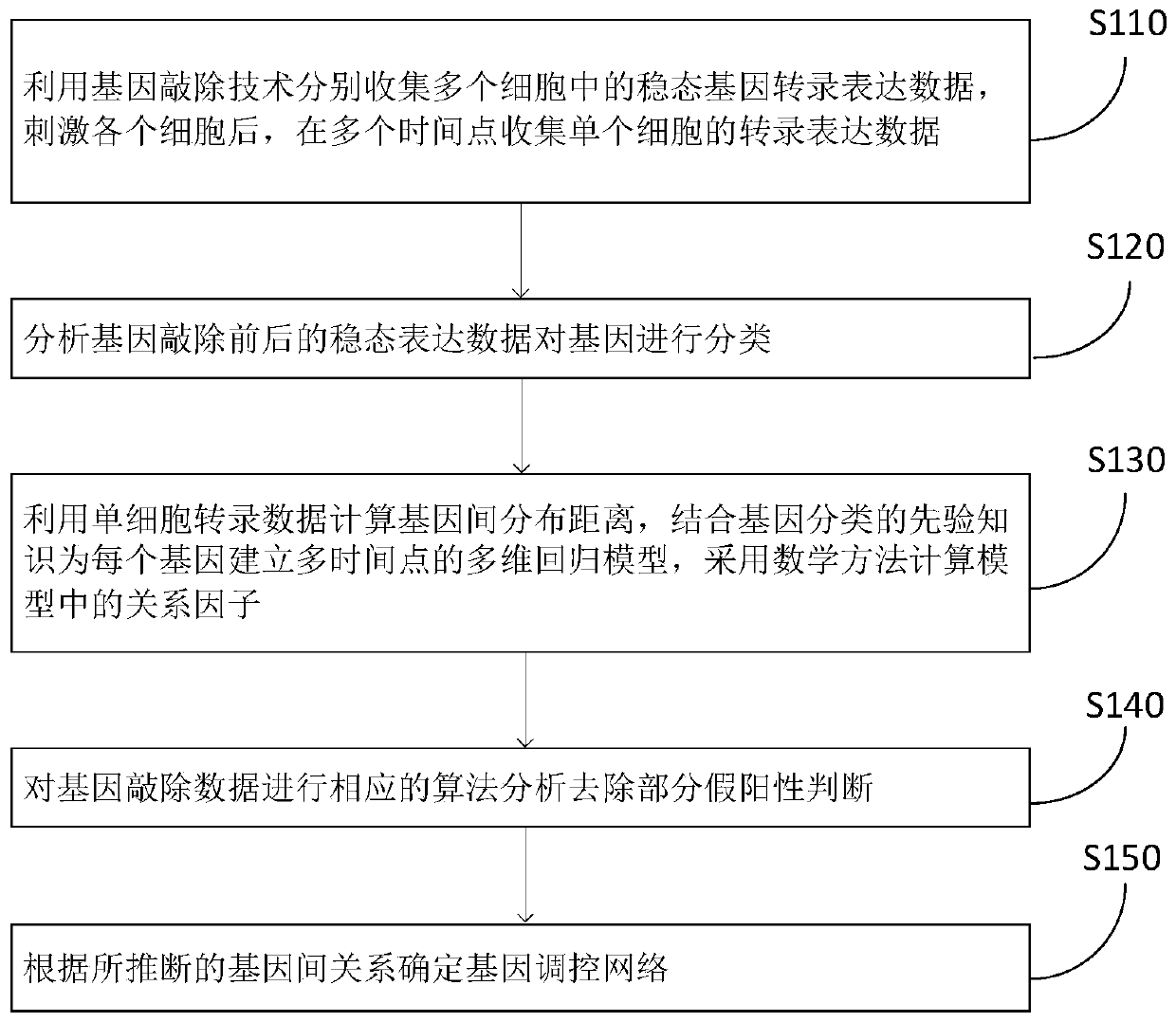
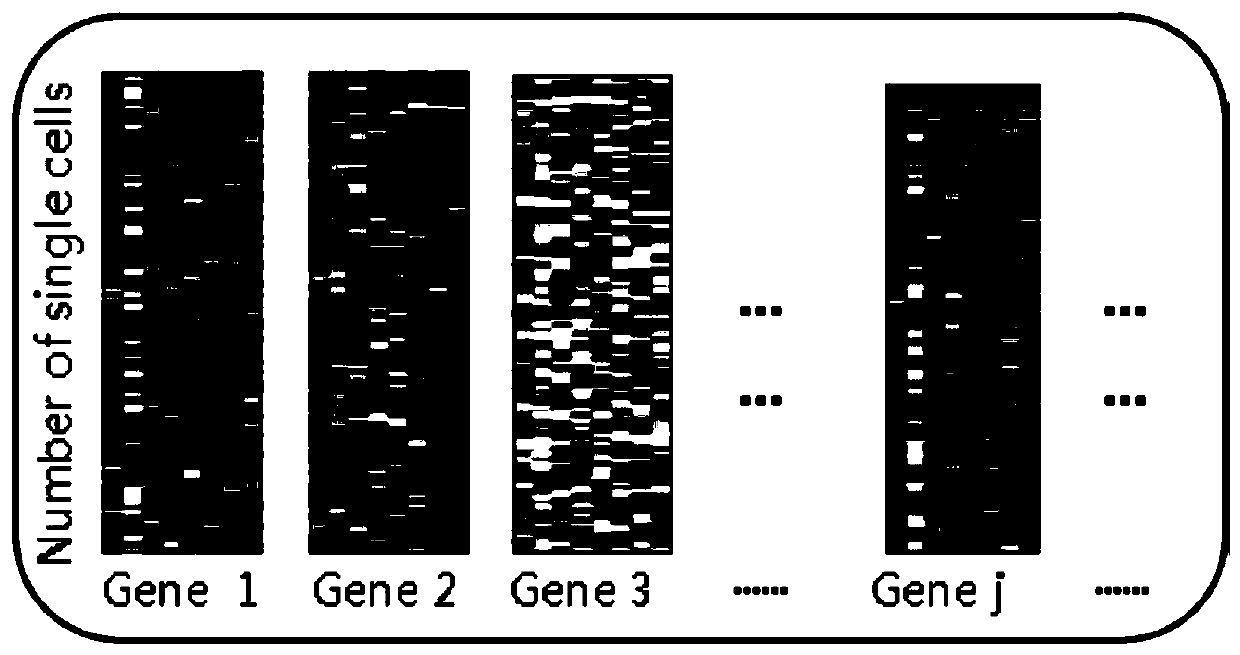
Method for deducing gene regulation network by using single cell transcription and gene knockout data
ActiveCN110517724AImprove accuracySolve problems with high computational complexityBiostatisticsSystems biologyComputation complexityMulti dimensional
The invention discloses a method for deducing a gene regulation network by using single cellular transcription and gene knockout data, which comprises the following steps of: classifying genes by analyzing steady-state expression data before and after gene knockout to serve as priori knowledge so as to reduce the space-time complexity and improve the deduction accuracy; calculating an inter-gene distribution distance by utilizing single cell transcription data, establishing a multi-time-point multi-dimensional regression model for each gene according to the gene classification result, and calculating a relation factor in the model by adopting a mathematical method; performing corresponding algorithm analysis on the gene knockout data to remove part of false positive judgment, and making upfor the defect of analyzing dynamic data. According to the method, the problem of high computation complexity in analyzing the time sequence single-cell data is effectively solved, and the accuracy of deducing the gene regulation network is improved.
Owner:TAIYUAN UNIV OF TECH
Method of determining relationships of gene expression and methylation modification regulation of predetermined species
ActiveCN108509769AImprove accuracyGood repeatabilityHybridisationSpecial data processing applicationsCandidate Gene Association StudyCorrelation analysis
The invention discloses a method of determining relationships of gene expression and methylation modification regulation of a predetermined species. The method comprises: (1) using a male parent and afemale parent of the predetermined species and a progeny sample thereof as to-be-tested samples, and carrying out bisulfite whole-genome methylation sequencing; (2) determining all cytosine sites inthe progeny sample and reads comprising the cytosine sites; (3) determining reads which are in the reads comprising the cytosine sites and and belong to allele sequences; (4) determining an apparent genotype of each pair of target reads; (5) carrying out gene classification on the target fragments; (6) carrying out counting of the number of target fragments comprised by each gene and the ratio ofthree apparent genotypes, and obtaining expression level information of each gene; (7) obtaining various candidate gene combinations by division; (8) carrying out Pearson correlation analysis; (9) screening out a target gene combination; and (10) carrying out multiple linear regression. Therefore, the relationships of gene expression and methylation modification regulation of the predetermined species can be effectively determined.
Owner:BEIJING FORESTRY UNIVERSITY
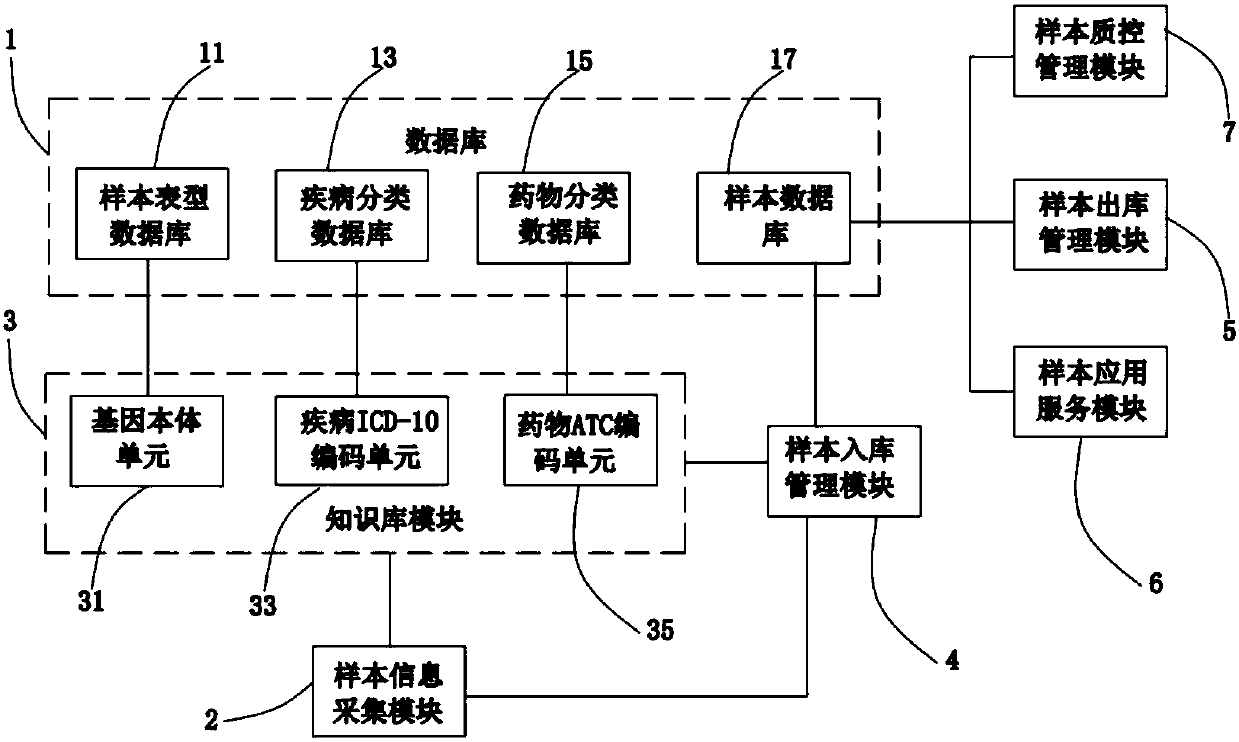
Bio-sample library management system suitable for pharmacogenomics
InactiveCN107871061AImprove scientific research efficiencyMeet needsProteomicsGenomicsDiseaseGenomics
The invention discloses a bio-sample library management system suitable for pharmacogenomics. The system comprises a database, a sample information collection module, a knowledge base module, a samplestorage management module, a sample output management module and a sample application service module, wherein the knowledge base module is in communication connection with the database, the sample information collection module and the sample storage management module; the sample storage management module is in communication connection with the database and the sample information collection module; and the sample output management module and the sample application service module are in communication connection with the database. According to the bio-sample library management system suitable for the pharmacogenomics, provided by the invention, sample management taking drug classification or gene classification as a center can be realized; and during sample application service, the demands of clinical pharmacogenomics can be better met and a drug use patient sample can be accurately output in combination with disease information to improve scientific and research efficiency.
Owner:CHANGSHA 3G BIOTECH
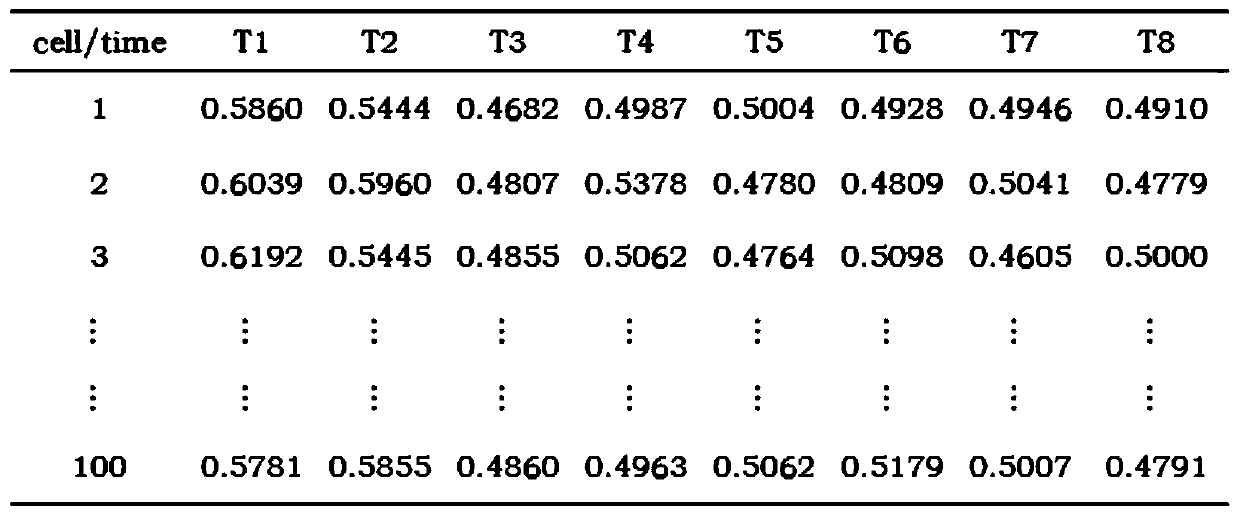
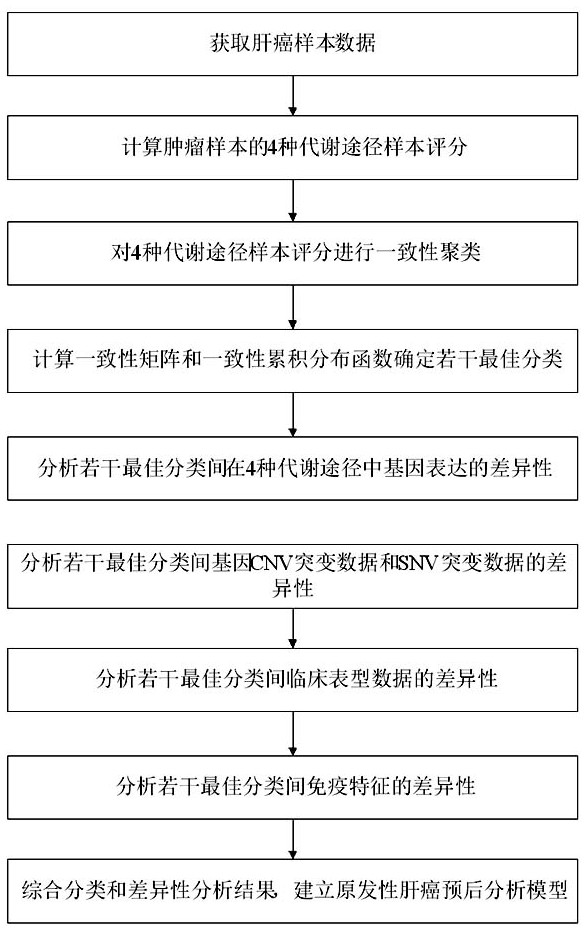
Primary liver cancer gene classification and liver cancer tissue energy metabolism-based prognosis analysis method
PendingCN114822688AAccurate prediction of prognosisBiostatisticsProteomicsTumor SampleClinical phenotype
The invention discloses a prognosis analysis method based on primary liver cancer gene classification and liver cancer tissue energy metabolism. The prognosis analysis method comprises the following steps: acquiring clinical phenotype data, expression profile data and gene CNV and SNV mutation data of a liver cancer sample; calculating sample scores of four metabolic pathways of the tumor sample, and determining a plurality of optimal classifications; and analyzing differences of expression, gene mutation, clinical phenotypes and immune characteristics of a plurality of optimal classification metabolic pathways, and then comprehensively analyzing results to establish a primary liver cancer prognosis analysis model. The method has the advantages that the primary liver cancer gene classifier is constructed through a big data analysis technology, the molecular subtype of the primary liver cancer is determined, and the metabolic pathway expression difference, the gene mutation difference, the clinical manifestation difference and the immune difference of the molecular subtype of the primary liver cancer are analyzed, so that the primary liver cancer prognosis analysis model is established; the method is used for accurately predicting the prognosis of the primary liver cancer and provides a research basis for revealing an energy metabolism mode and a tumor microenvironment of the primary liver cancer.
Owner:THE FIRST AFFILIATED HOSPITAL ZHEJIANG UNIV COLLEGE OF MEDICINE
Biological sequence processing and model training method
PendingCN114881131AScale upImprove performanceCharacter and pattern recognitionSequence analysisData setData mining
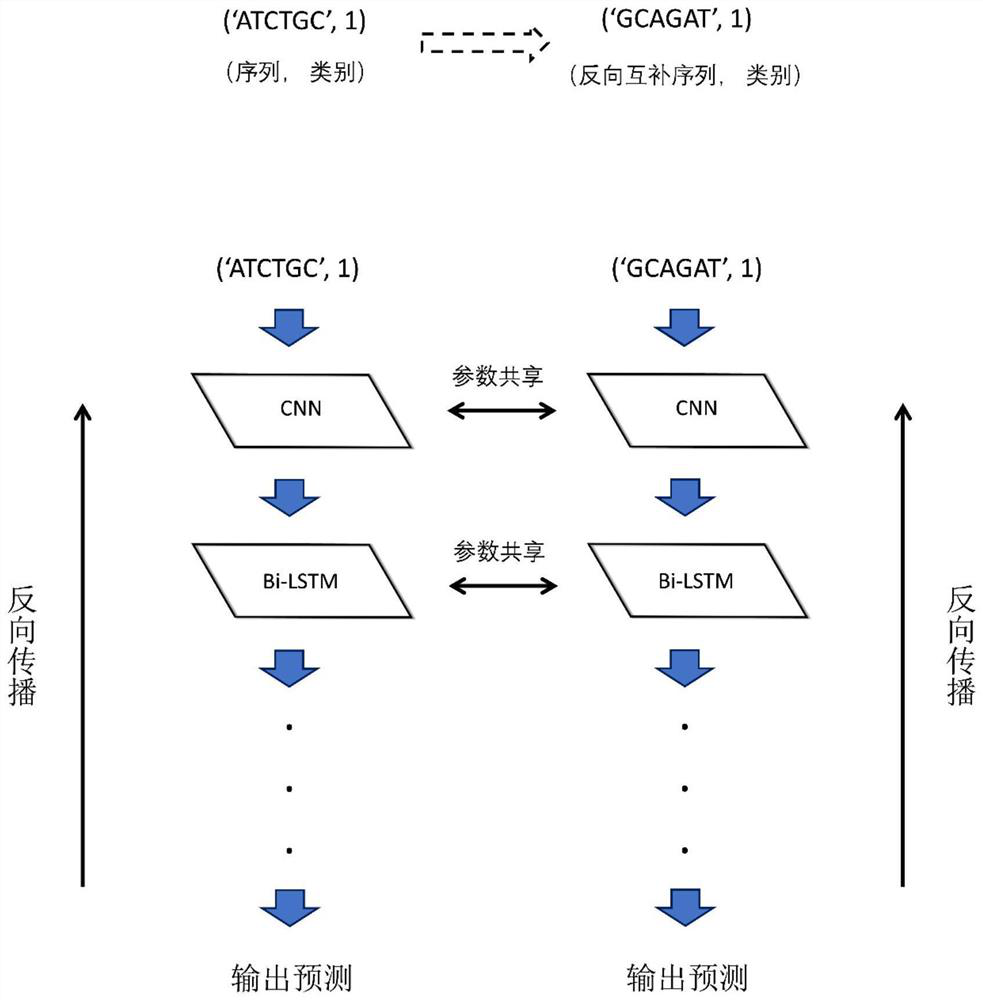
The invention provides a biological sequence processing and model training method. The method comprises the following steps: S1, acquiring data of a biological gene sequence and integrating the data; s2, preprocessing the data, traversing the read biological gene sequences, and filtering out biological gene sequences meeting requirements; s3, constructing a data set required by the training model, and finely adjusting the data set according to the number of each category of data in the data set to ensure that the scales of the various categories of data in the data set are approximately equal; s4, carrying out quantity balance and gene data length balance processing on the data of the data set to obtain a training set; and S5, training a model with a reverse complementary network by using the training set. The method provided by the invention can save time on the basis that the accuracy of the method is similar to that of a traditional gene classification and recognition method, and can correctly predict part of genes which cannot be correctly classified by a traditional biological method.
Owner:ZHEJIANG UNIV CITY COLLEGE
Gene classification method and related equipment
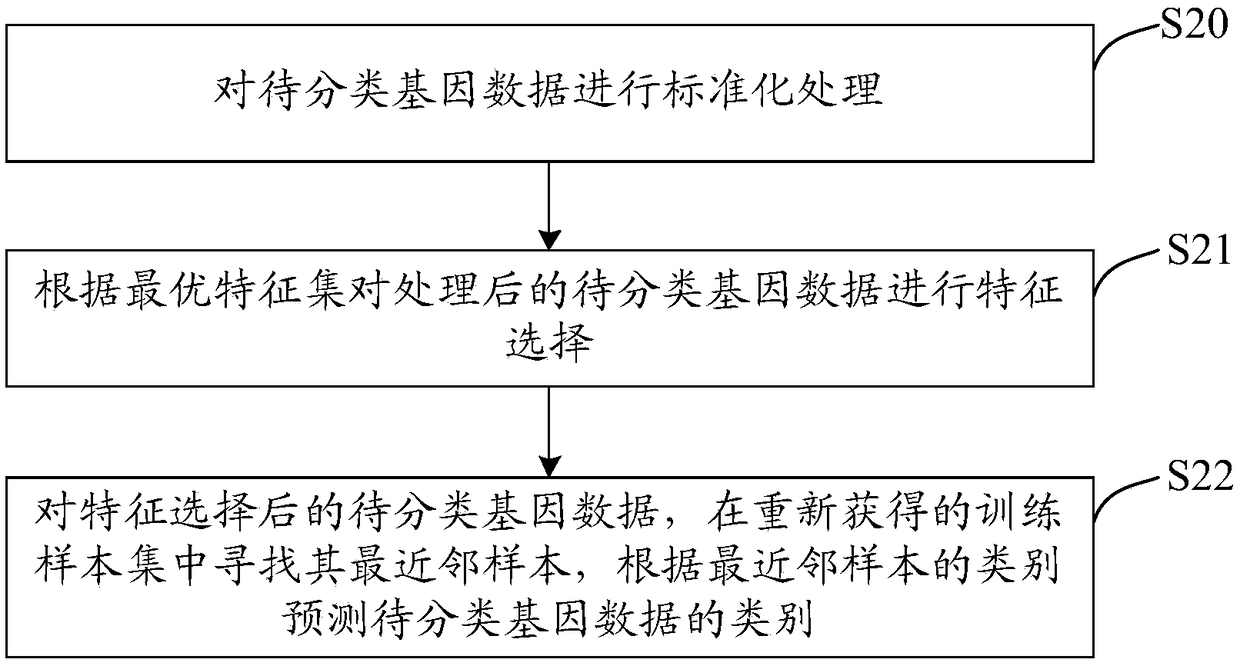
InactiveCN108763873AWith sparsenessThere is sparsitySpecial data processing applicationsFeature setAlgorithm
The invention discloses a gene classification method and related equipment. The method comprises the steps of carrying out standardization processing on input gene data serving as training samples, wherein the gene data comprises a plurality of attributes; by difference values between intervals between the training samples and heterogeneous neighbor samples and intervals between the training samples and similar neighbor samples, and weight vectors of the corresponding attributes, building a logical regression optimization function which represents expected interval difference of all the training samples, building a minimized optimization model by taking the weight vectors of the corresponding attributes as norm constraint terms, and performing calculation through iterative operation to obtain the weight vectors of the corresponding attributes; and sorting attribute features according to values of the obtained weight vectors, carrying out classification training on the gene data servingas the training samples according to the sorted attribute features, and obtaining an optimal feature set as a classification basis, thereby classifying the to-be-classified gene data. According to the gene classification method and the related equipment provided by the invention, the relatively high classification precision can be obtained.
Owner:SUZHOU UNIV
Gene classification method and device
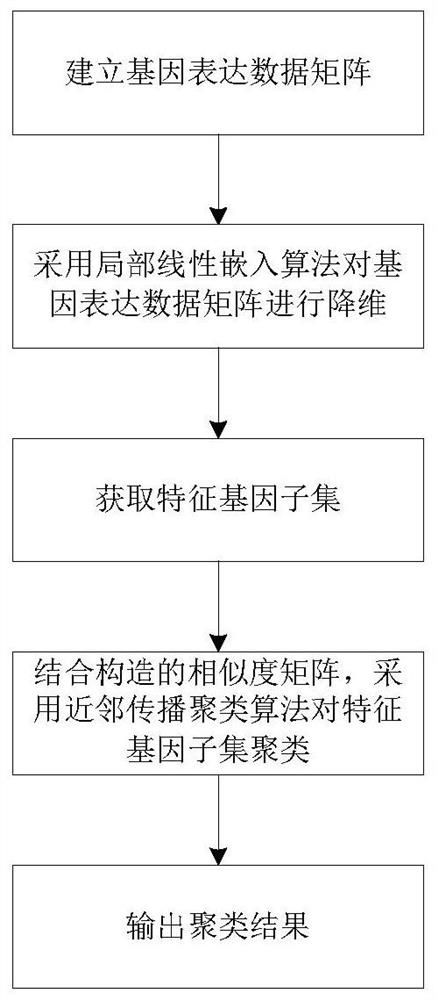
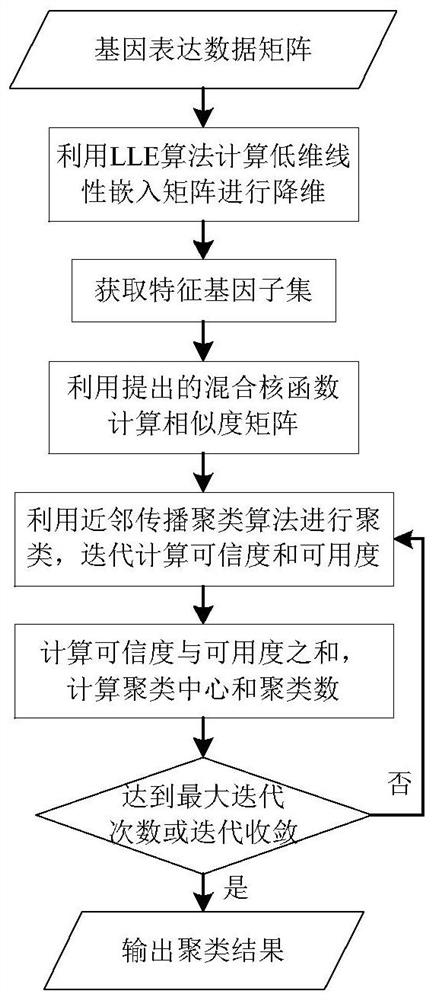
ActiveCN108171012BThe clustering result is accurateImprove clustering effectBiostatisticsHybridisationCluster algorithmAlgorithm
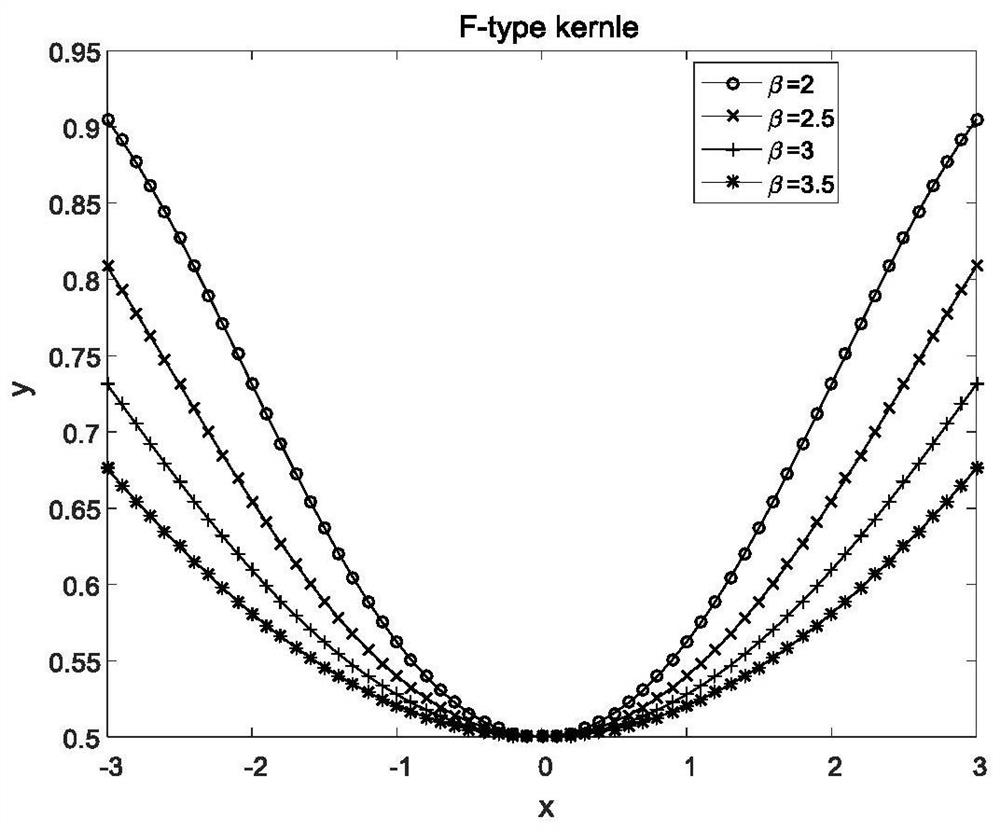
The invention relates to a gene classification method and device, which combines the LLE algorithm and the AP clustering algorithm, and uses the proposed mixed kernel function to improve the similarity measurement function. First, the LLE algorithm is used to map the original high-dimensional gene expression data set to a low-dimensional space to achieve the purpose of dimensionality reduction; secondly, a new global kernel function is proposed as the F-type kernel function, and it is linearly combined with the Gaussian kernel function to form a new The hybrid kernel function, and use the proposed hybrid kernel function to calculate the similarity measure, construct a new similarity matrix S; then cluster the data through the AP clustering algorithm and the similarity matrix, iteratively obtain the final clustering result; finally through Compared with other clustering methods, the validity and accuracy of the algorithm of the present invention are verified.
Owner:HENAN NORMAL UNIV
A kind of human leukocyte hla-b*27 genotyping primer set and application
ActiveCN109182491BIncrease motivationGuaranteed reliabilityMicrobiological testing/measurementDNA/RNA fragmentationWhite blood cellMedicine
The invention discloses a human leukocyte HLA‑B*27 genotyping primer set, which includes detecting blood types A and A respectively. 205 , B and O (261G缺失) , O 1 , O 2 The allelic primer pair AR and AF; A 2 R and A 2 F;BR and BF;0 T R and O T F;O 1 R and O 1 F;O 2 R and O 2 F and internal reference primer pair; the allelic primer set designed by the present invention has high power and strong specificity, and the internal reference primer pair internal reference 1R and internal reference 1F, internal reference 2R and internal reference 2F are designed according to the conserved fragment of human albumin gene, and each detection The number of pairs of allelic primers in the well is 2 to 4 pairs, and at the same time, it contains 2 pairs of internal references to ensure the quality and correctness of PCR amplification, thereby ensuring the reliability of the test results.
Owner:江苏中济万泰生物医药有限公司
A genotyping detection kit for 23 gene loci of human rh blood type
ActiveCN108410972BAmplified equalizationIncrease mutual interferenceMicrobiological testing/measurementGroup A - bloodGenotyping
The invention belongs to the technical field of in vitro detection of human nucleic acid, and particularly relates to a genotyping detection kit for human No. 5, No. 19, No. 21 and 23 gene loci of X / Y chromosomes. The present invention first designs a compound PCR amplification system for genotyping 23 gene loci distributed on human chromosomes 5, 19, 21 and X / Y at the same time, wherein the PCR amplification primer set is: SEQ ID No.1 to SEQ ID No.53; the genotyping detection kit of the present invention includes a primer composition container and a PCR reaction mother solution container; the primer composition container contains the primer composition of SEQ ID No.1 to SEQ ID No.53 stock solution. The invention realizes single-tube amplification of 23 gene loci, the gene loci in each fluorescence channel and in different fluorescence channels are amplified evenly, and the typing map is good; and the cost, manpower and time can be significantly saved, and work is improved. efficiency.
Owner:上海五色石医学科技有限公司
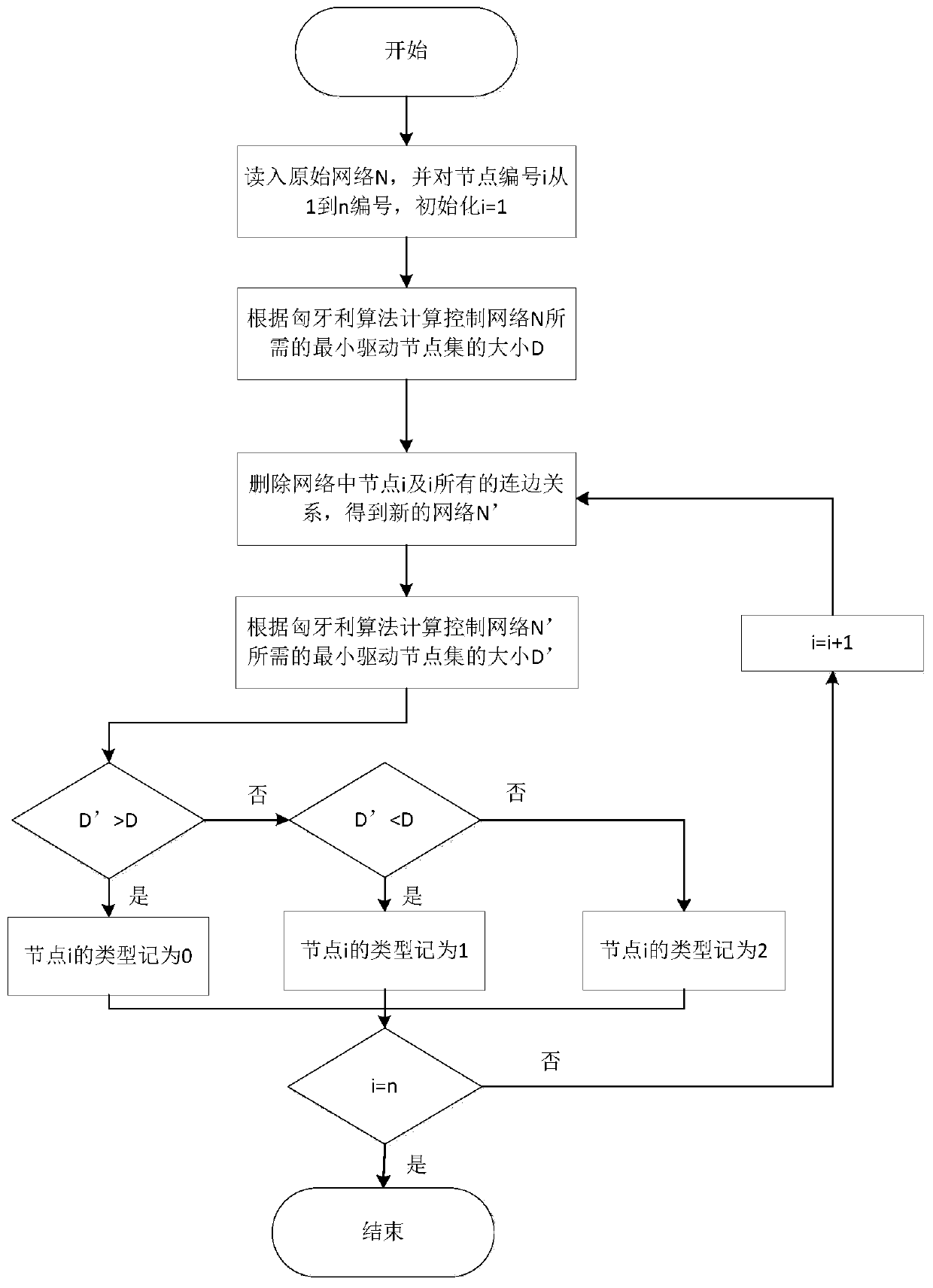
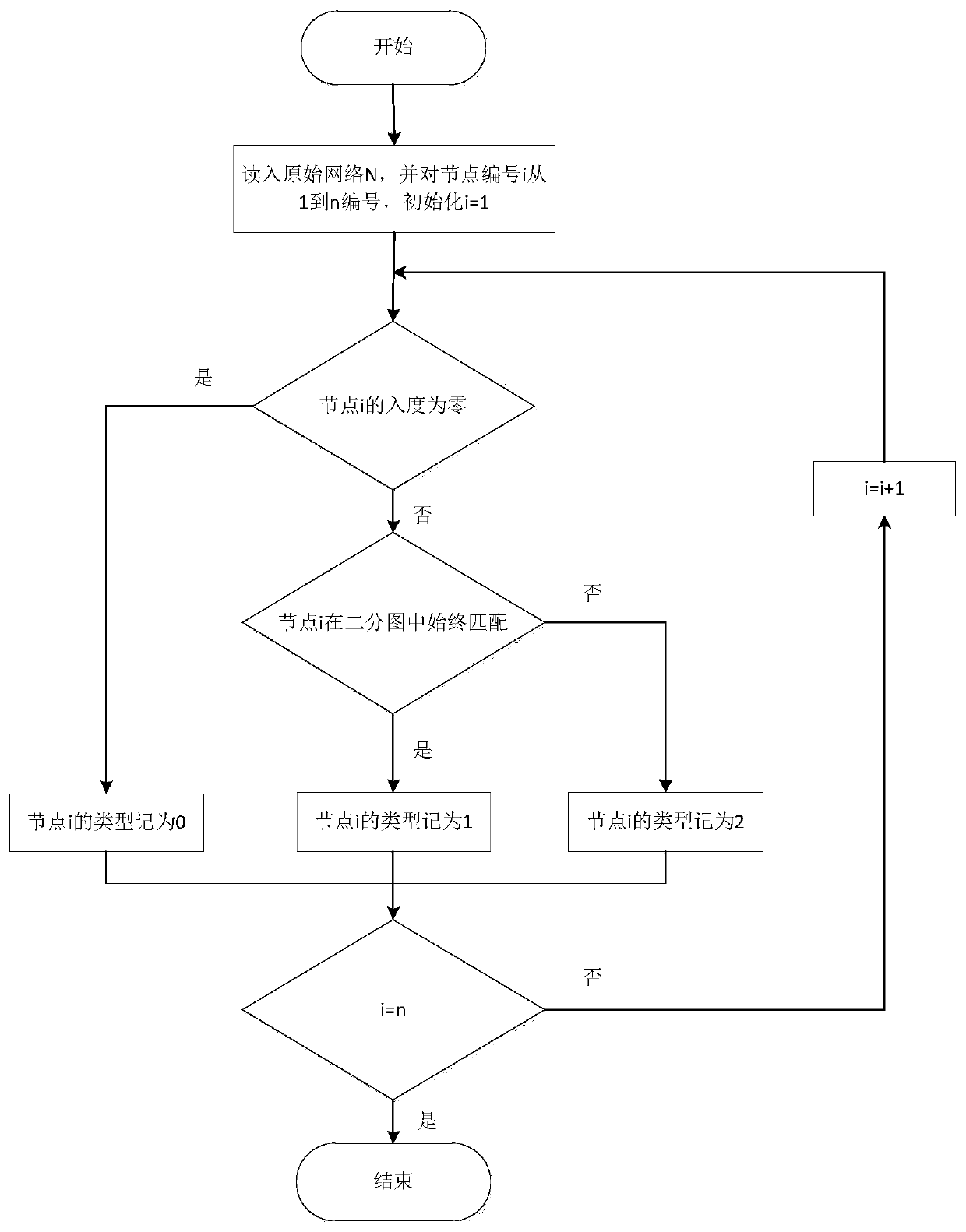
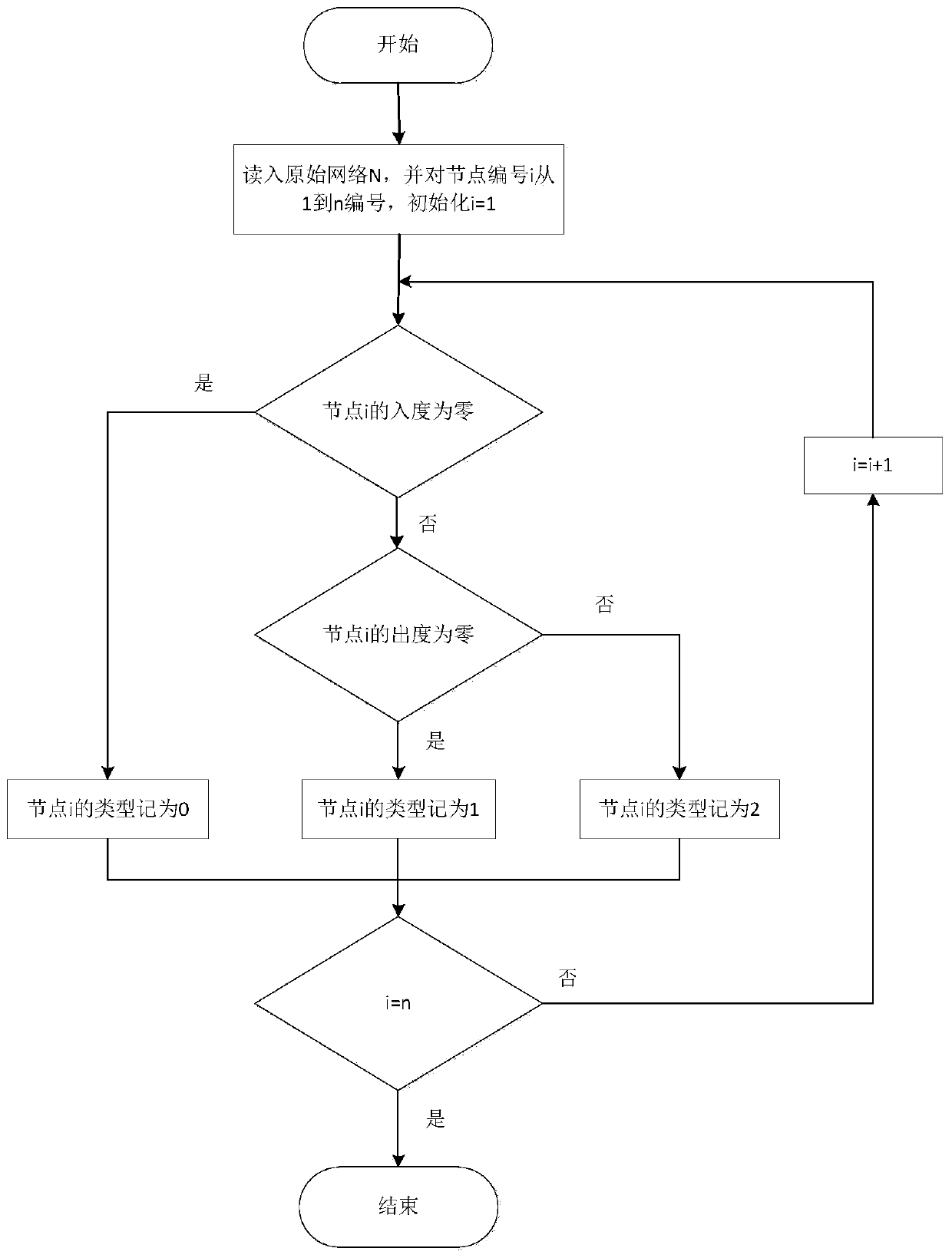
Method for identifying controllable gene based on complex network structure
The invention provides a method for identifying controllable gene based on complex network structure. A controllability node classification framework is constructed; genes are divided into different types for controlling role differences; a new gene is identified through statistical significance. According to the gene recognition method based on the controllable node classification framework, global information in a network is considered, gene classification is achieved from multiple control levels, the framework is applied to a tissue-specific regulation and control network, genes with remarkable biological significance can be systematically detected, and a tool platform is provided for further gene research.
Owner:XIAN UNIV OF TECH
Cancer gene classification method and device based on two-stage depth feature selection and storage medium
ActiveCN112926640AAvoid not being selectedEasy to identifyCharacter and pattern recognitionNeural learning methodsCancer geneBiology
The invention relates to a cancer gene classification method and device based on two-stage depth feature selection and a storage medium. The method comprises the steps: A, training a cancer gene classification model: (1) obtaining training data: in the first stage, integrating three feature selection algorithms to carry out comprehensive feature selection, and obtaining a feature subset; in the second stage, the optimal representation of a feature subset is obtained by using an unsupervised neural network; (2) dividing the optimal representation of the feature subset into a training set and a test set, and inputting the training set and the test set into a neural network for training; and B, cancer gene classification: preprocessing to-be-detected cancer gene data, and inputting the preprocessed to-be-detected cancer gene data into the trained cancer gene classification model to realize cancer gene classification. According to the invention, by using the integrated feature selection method, feature selection is carried out in consideration of all aspects; and the optimal representation of the features is extracted by using the unsupervised neural network, so that cleaner gene features are obtained, and the classification precision is improved.
Owner:QILU UNIV OF TECH
A method for identifying key tumor genes based on particle swarm optimization and scoring criteria
ActiveCN106951728BAccurate identificationEffectively deleteProteomicsGenomicsGene recognitionParticle swarm algorithm
Owner:JIANGSU UNIV
A distance measurement method for gene expression profiles based on deep learning
ActiveCN110033041BAccurate and fast calculationCharacter and pattern recognitionNeural architecturesGene FeatureExpression analysis
The invention belongs to the field of gene expression spectrum classification, discloses a gene expression spectrum distance measurement method based on deep learning, and belongs to the mining and application of deep learning on biological big data. First, a convolutional neural network model suitable for gene feature metric learning is designed to extract the characteristics of the data, and then the distance between the data is calculated by using the improved cosine distance, and finally the classification effect of the classification algorithm is used to measure the performance of the method excellent. This method can quickly and efficiently measure the similarity between different gene expression profiles, and provide data for subsequent studies such as gene classification, clustering, differential expression analysis, and compound screening. Compared with the traditional gene enrichment method, this method significantly improves the distance measurement effect between data, and can effectively reduce the manual intervention in gene expression profile analysis, avoiding the over-fitting phenomenon that is easy to occur in conventional deep networks. This method has strong transferability.
Owner:HUNAN UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com