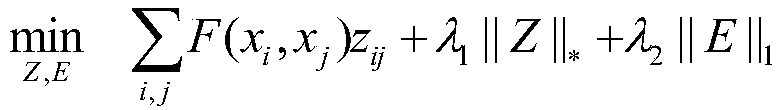Method of identifying cell types based on single-cell RNA sequencing data
A technology for sequencing data and single cells, applied in the cross-research field of mathematics and biology, can solve problems such as the need to improve accuracy and efficiency, and achieve the effect of improving the clustering effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0056] The beneficial effects of the present invention will be described in detail below in conjunction with examples, aiming to help readers better understand the essence of the present invention, but not to limit the implementation and protection scope of the present invention.
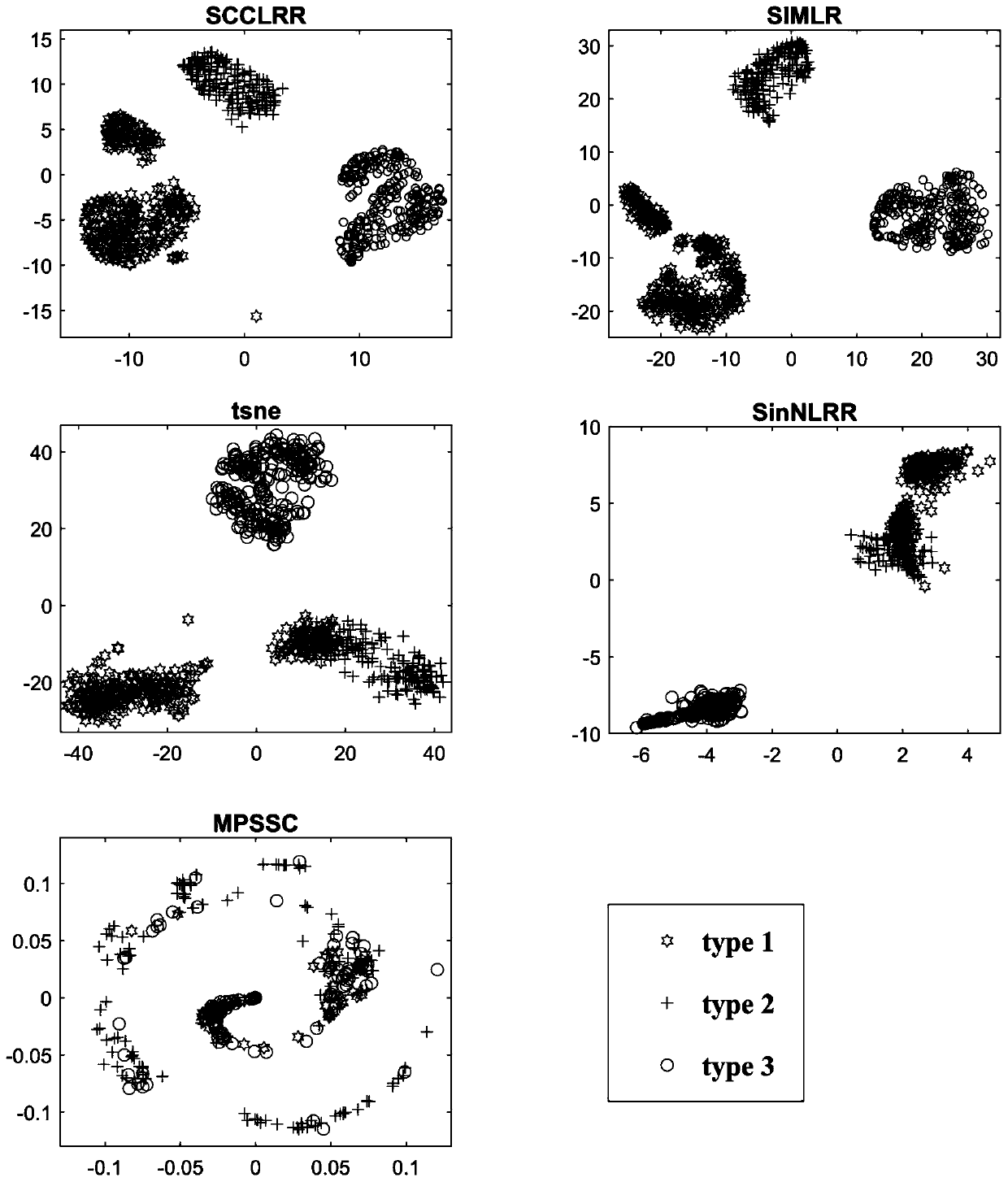
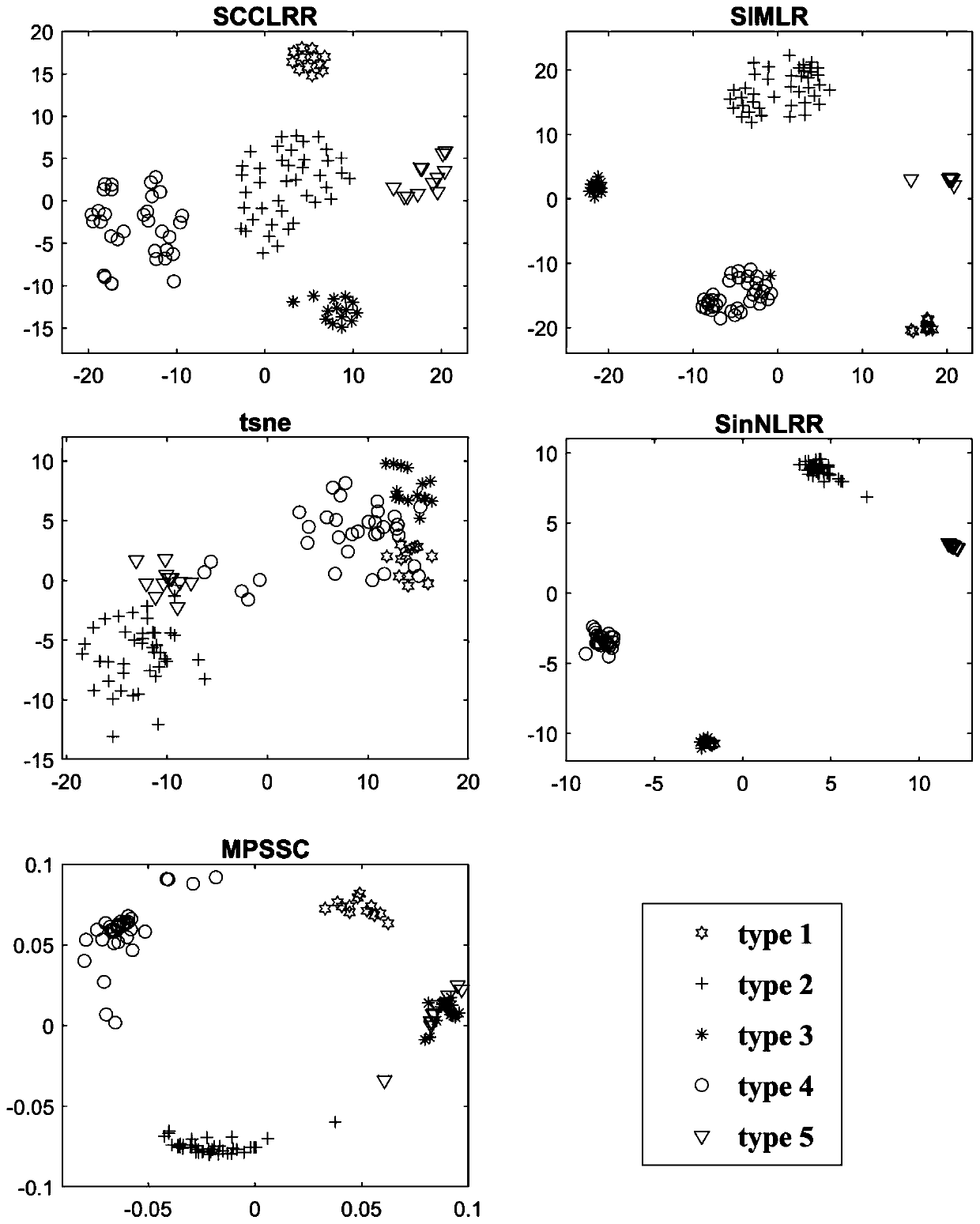
[0057] The present invention provides a method for identifying cell types based on single-cell RNA sequencing data, which is based on a matrix low-rank representation model and a method of graph regularization constraints to cluster noisy high-dimensional sparse single-cell RNA sequencing data for effective mining Global structural features and local association properties from single-cell RNA-sequencing data, leading to new computational methods for predicting key proteins. The main steps of the method include:
[0058] (1) Based on the single-cell RNA sequencing data X, the construction of the similarity matrix between cells is transformed into an optimization problem, and the mathematical model o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com