Liver cancer tumor marker heat shock factor 2 binding protein and application thereof
A tumor marker and binding protein technology, which is applied in the field of heat shock factor 2 binding protein, a tumor marker of liver cancer, can solve the problems of no published research on malignant tumors and less HSF2BP.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
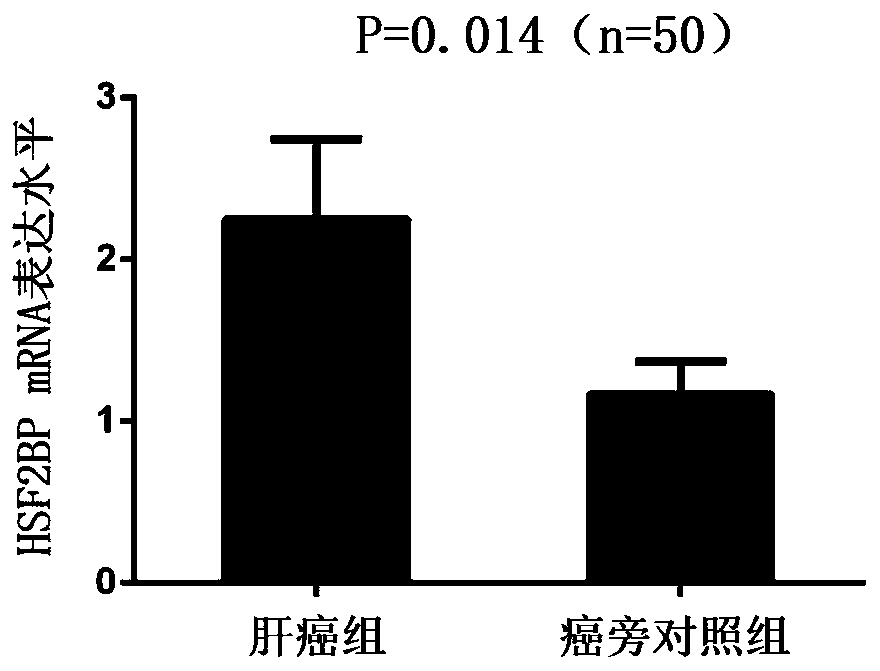
[0035] Example 1: Real-time quantitative PCR detection of HSF2BP mRNA expression in liver cancer tissues and corresponding adjacent normal tissues.
[0036] Patients with primary liver cancer who were admitted to the First Affiliated Hospital of Xi'an Jiaotong University and underwent "hepatectomy" from October 2012 to December 2016 were randomly selected, and 99 pairs of hepatocellular carcinoma and adjacent normal liver tissues (at least Tumor edge 3cm). The entire collection and experimental testing process was certified by the Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong University. Informed consent was signed for all tissue samples before collection. After the tissue specimens were obtained, they were immediately stored in a -80° refrigerator.
[0037] 1. Extraction of total cellular RNA using Trizol reagent
[0038] Materials and reagents: Trizol reagent, chloroform, isopropanol, DEPC water, 75% ethanol.
[0039] experiment procedure:
[0040]...
Embodiment 2
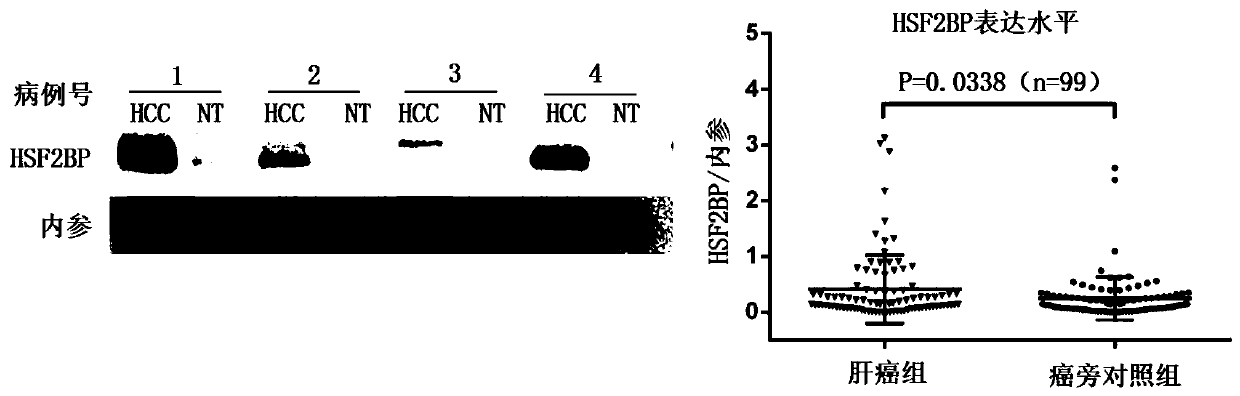
[0059] Example 2: Detection of the expression of HSF2BP protein in liver cancer tissue and corresponding adjacent normal tissue by Western Blot
[0060] 1. Tissue protein extraction process:
[0061] 1. Obtain 30mg of tissue protein and put it into a 2ml homogenization tube, add 300ul of pre-cooled RIPA lysate respectively;
[0062] 2. Add steel balls and crush for 1 minute at high speed on the tissue crusher;
[0063] 3. Take out the steel ball, place it on ice together with the mixing tube, and fully crack for 30 minutes;
[0064] 4. After centrifuging at low temperature and high speed for 15 minutes, carefully obtain the supernatant and transfer it to a new 1.5ml centrifuge tube;
[0065] 5. Take 10ul protein sample, use BCA protein quantification method to quantify, and obtain the concentration of sample protein;
[0066] 6. Add the remaining protein samples to 5×loading buffer in proportion and vortex to mix;
[0067] 7. Protein samples were heat-treated in boiling wa...
Embodiment 3
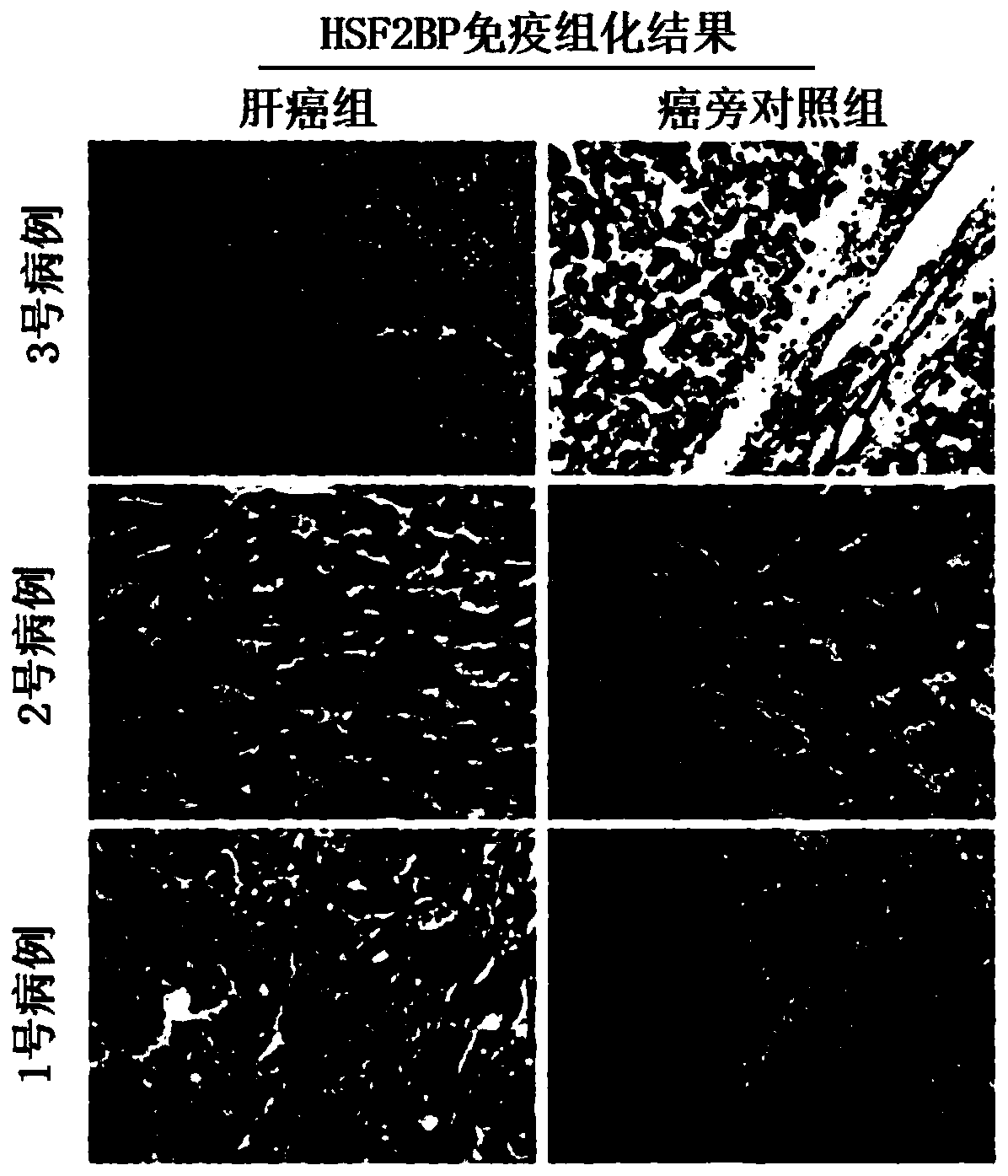
[0081] Example 3: Immunohistochemical method to detect the expression of HSF2BP protein in liver cancer tissues and corresponding normal adjacent tissues
[0082] 1. Paraffin tissue section
[0083] 1. Place the clinical specimen tissue in 4% paraformaldehyde solution and fix it at room temperature for 24 hours;
[0084] 2. Rinse with PBS 2-3 times, place the tissue in 80% alcohol for 48 hours, 95% alcohol for 12 hours, absolute alcohol for 2 hours, and xylene for 2 hours to dehydrate;
[0085] 3. Immerse the tissue in wax for 30 minutes;
[0086] 4. After routine embedding, slice serially with a thickness of 5um;
[0087] 5. Place the slices in a 37°C incubator for 48 hours and store at room temperature.
[0088] 2. Immunohistochemical staining
[0089] 1. Dewax the paraffin-embedded sections in xylene for 2 times, 10 min each time;
[0090] 2. Gradient alcohol dehydration;
[0091] 3. Place the slices in methanol solution of 0.3% hydrogen peroxide for 15 minutes at roo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com