Detecting somatic single nucleotide variants from cell-free nucleic acid with application to minimal residual disease monitoring
A cell nucleic acid, detection body technology, applied in the direction of microbial determination/inspection, genomics, instruments, etc., can solve the problems of simplicity, inability to consider the impure nature of cfDNA, and inability to consider overlapping read partners, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
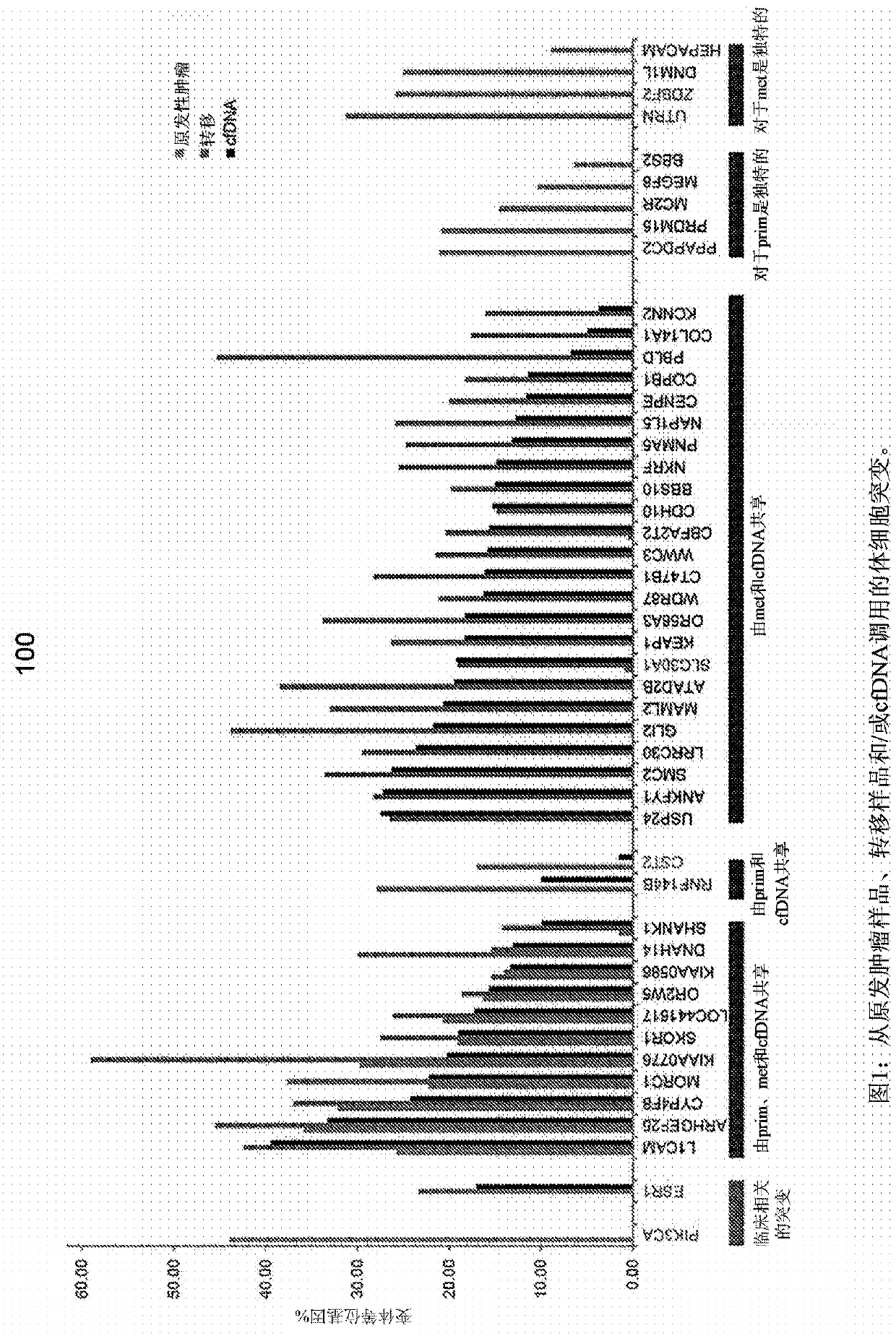
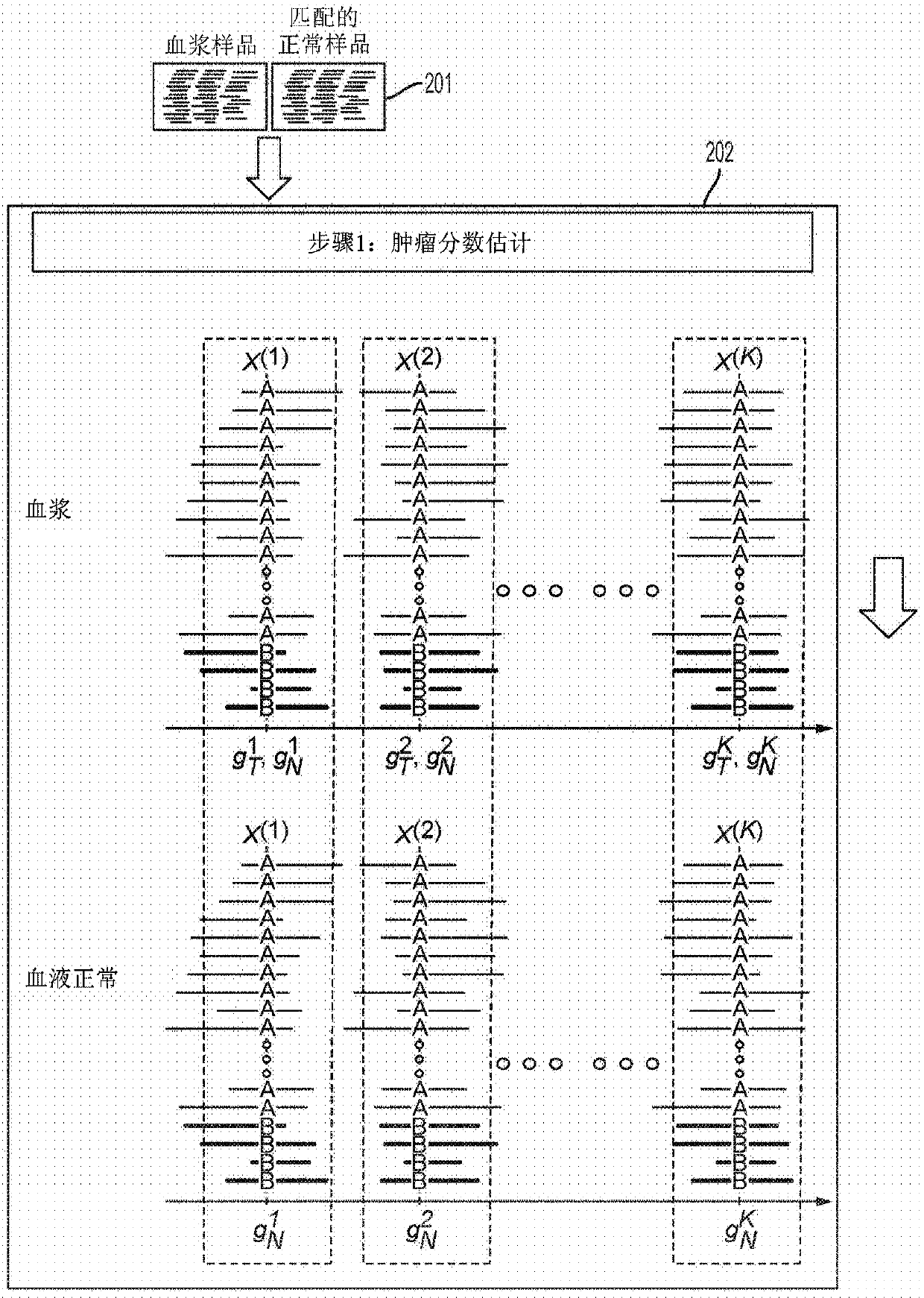
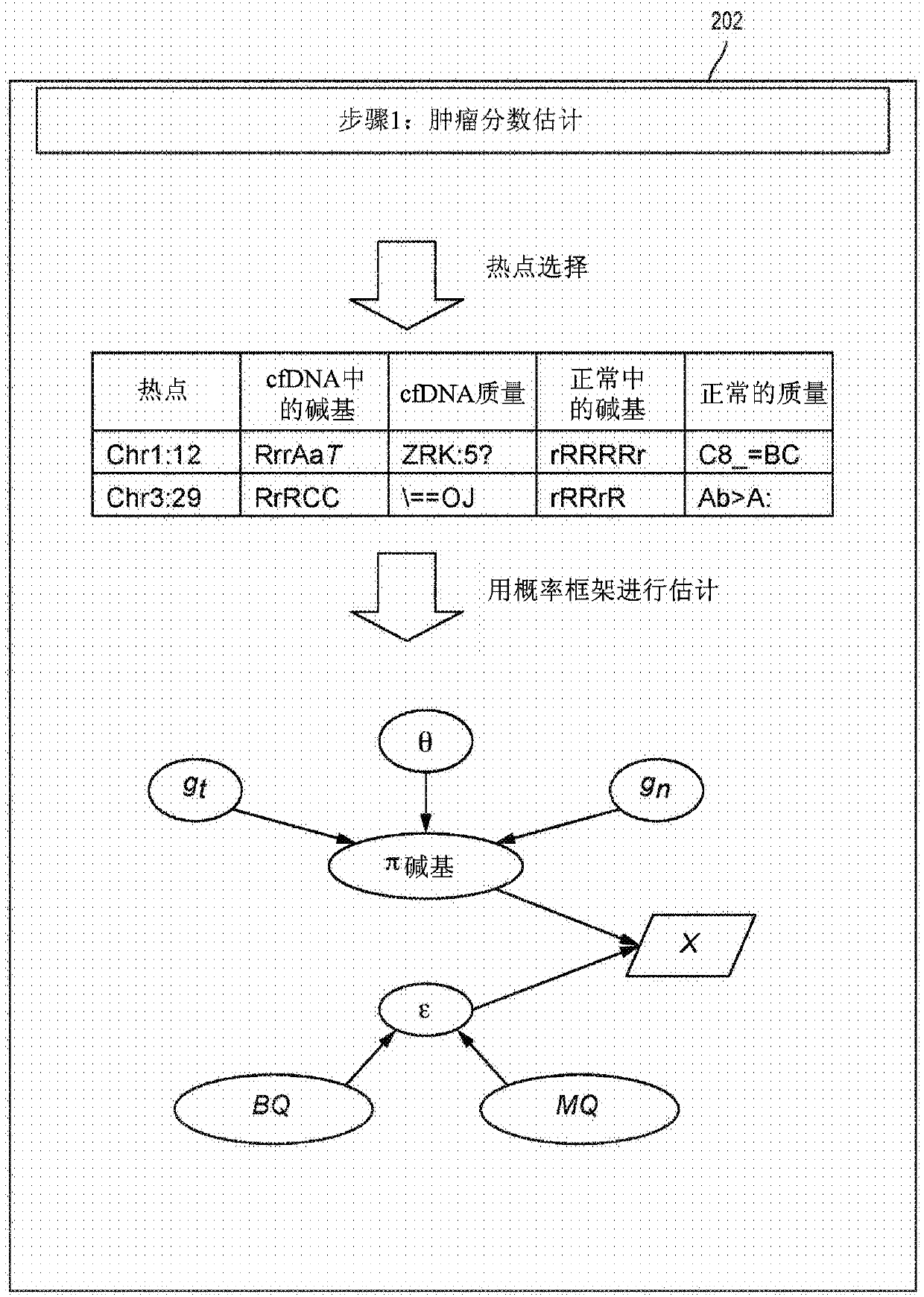
[0056] The present disclosure provides probabilistic models for accurate and sensitive somatic single nucleotide variant (SNV) detection in cell-free deoxyribonucleic acid (cfDNA) samples and / or cell-free ribonucleic acid (cfRNA) samples, The sample comprises a sequence data set. cfDNA samples can be taken from plasma samples. The model can receive as input sequencing data from cfDNA and matched normal samples, such as from white blood cells in blood. In addition, since joint genotypes can be determined for each locus in a sequence dataset, germline mutations can be removed. In addition, since the overall tumor cfDNA fraction can be taken into account, accurate SNV detection can be performed from samples with low tumor fractions that might otherwise be unavailable.
[0057] As described herein, cfDNA from a subject (eg, a cancer patient) can comprise a mixture of DNA or RNA from cells of a diseased organ (eg, tumor cells) and DNA or RNA from normal cells. Therefore, the gen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com