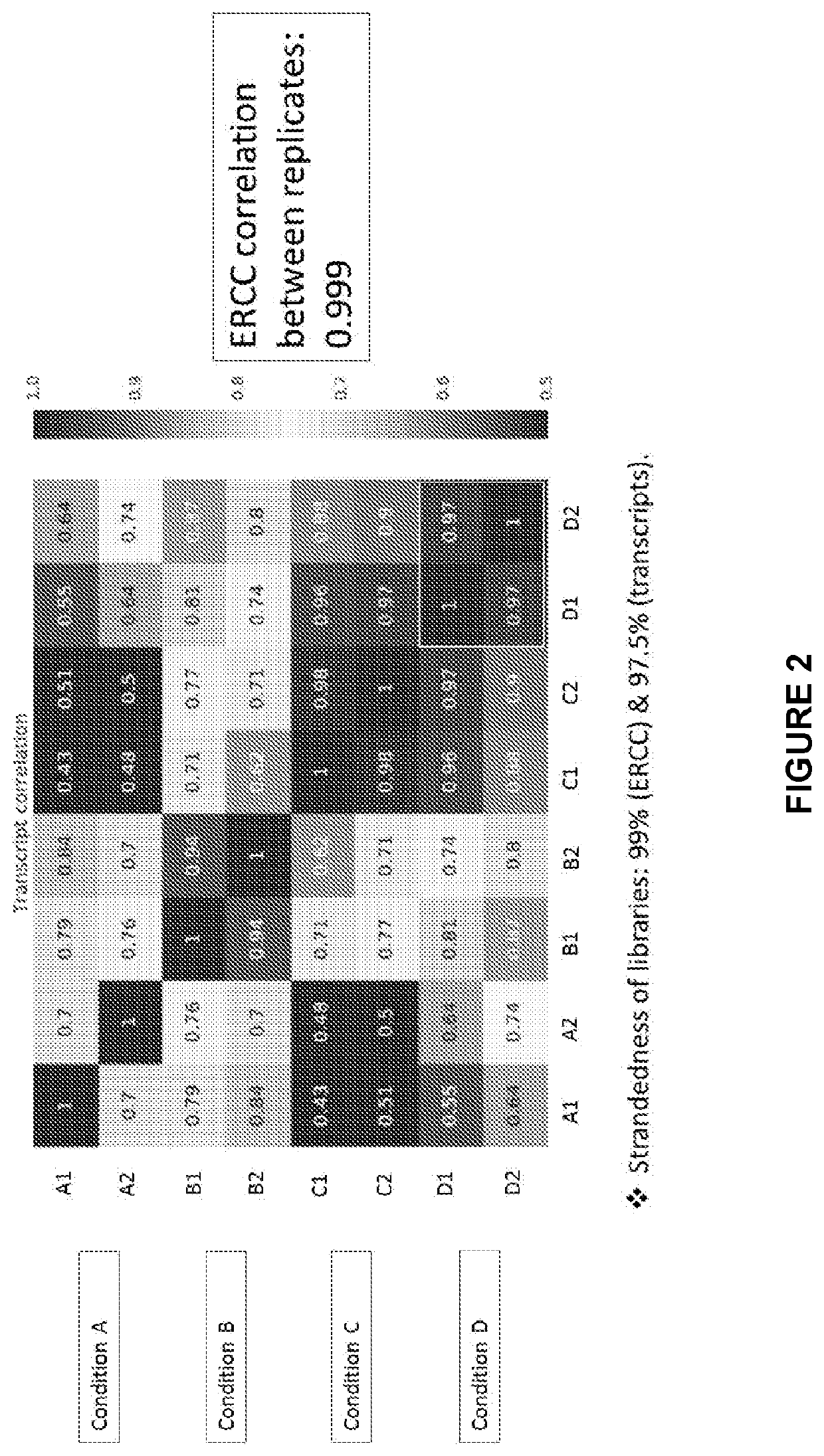
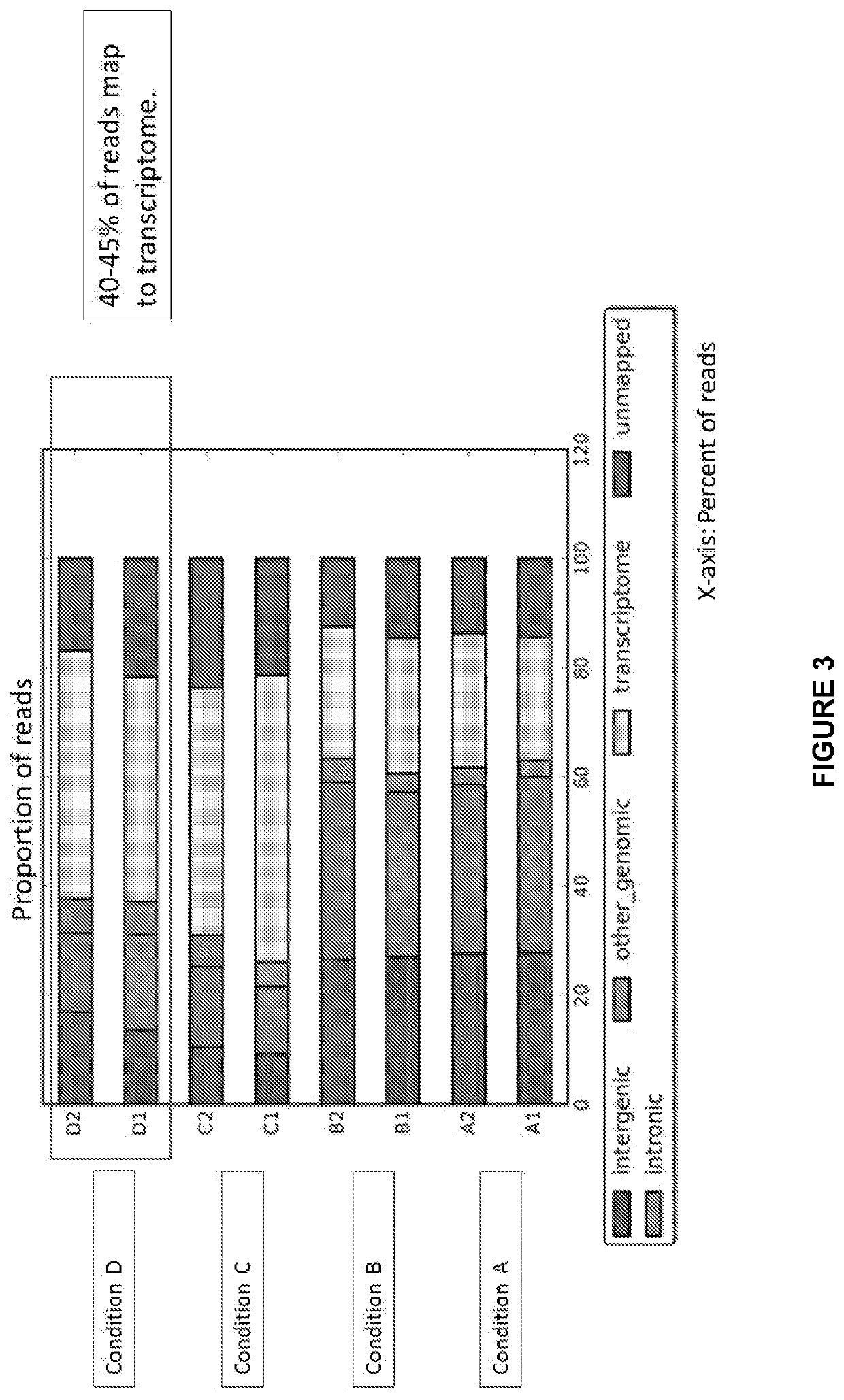
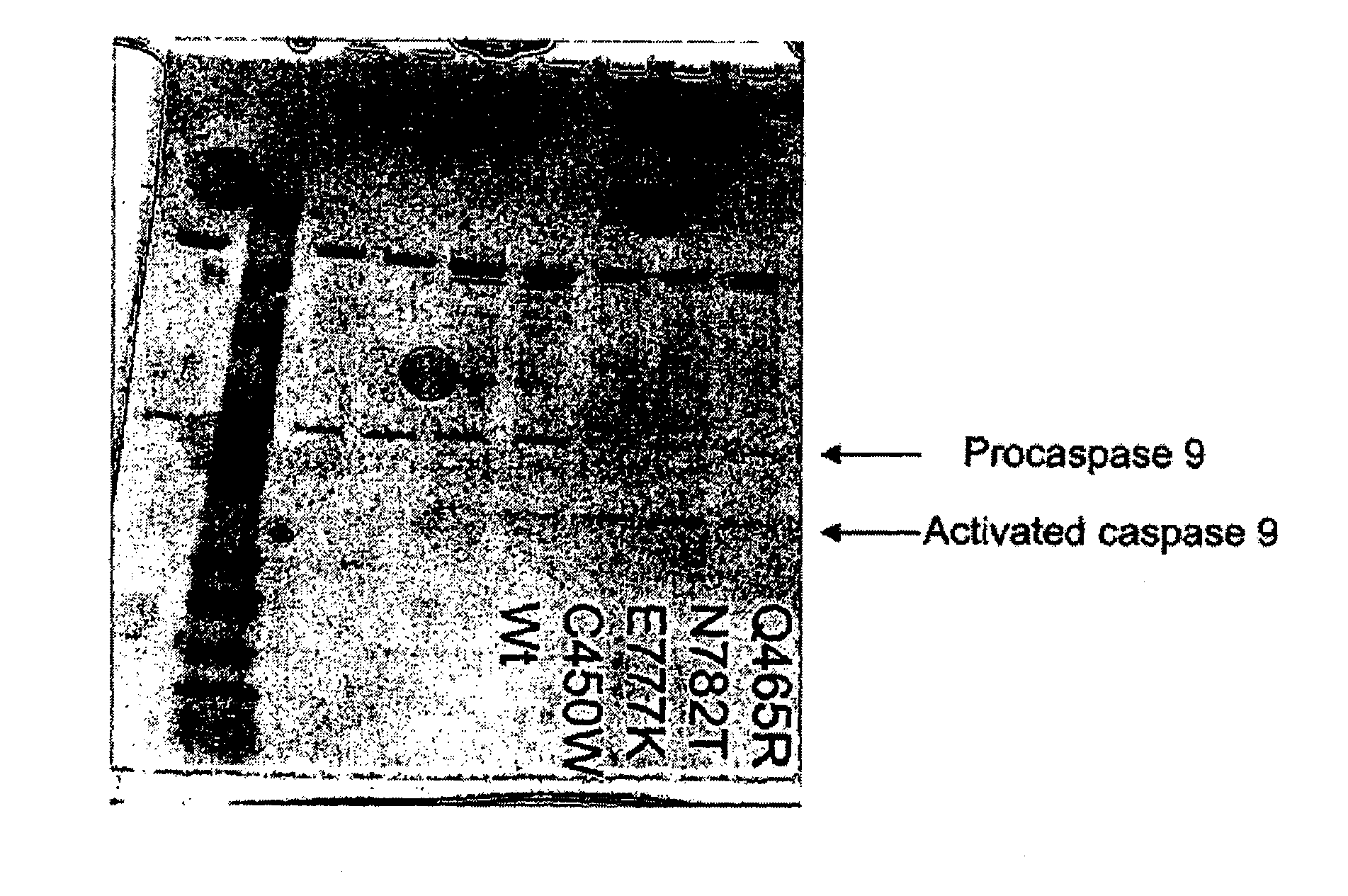
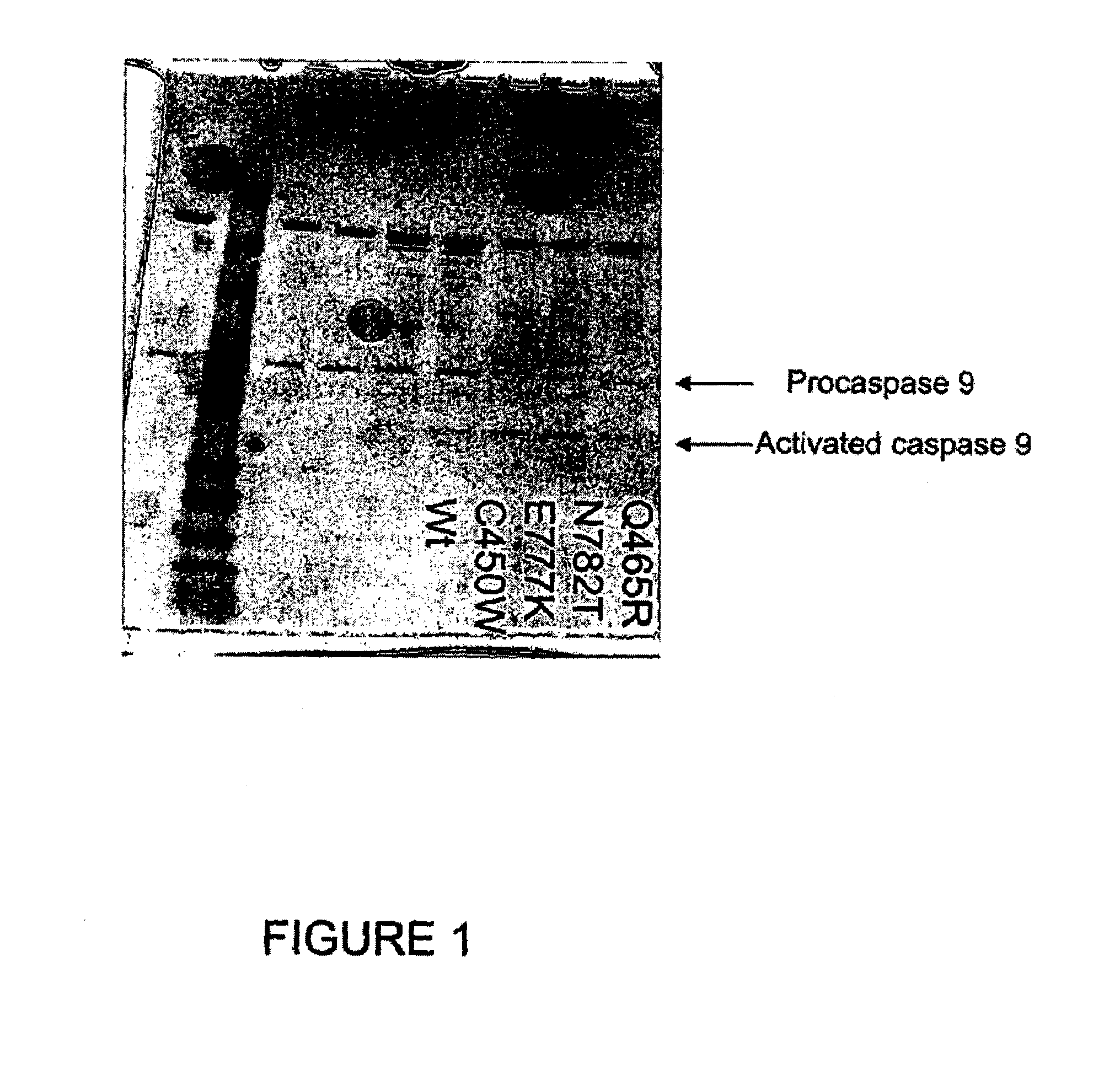
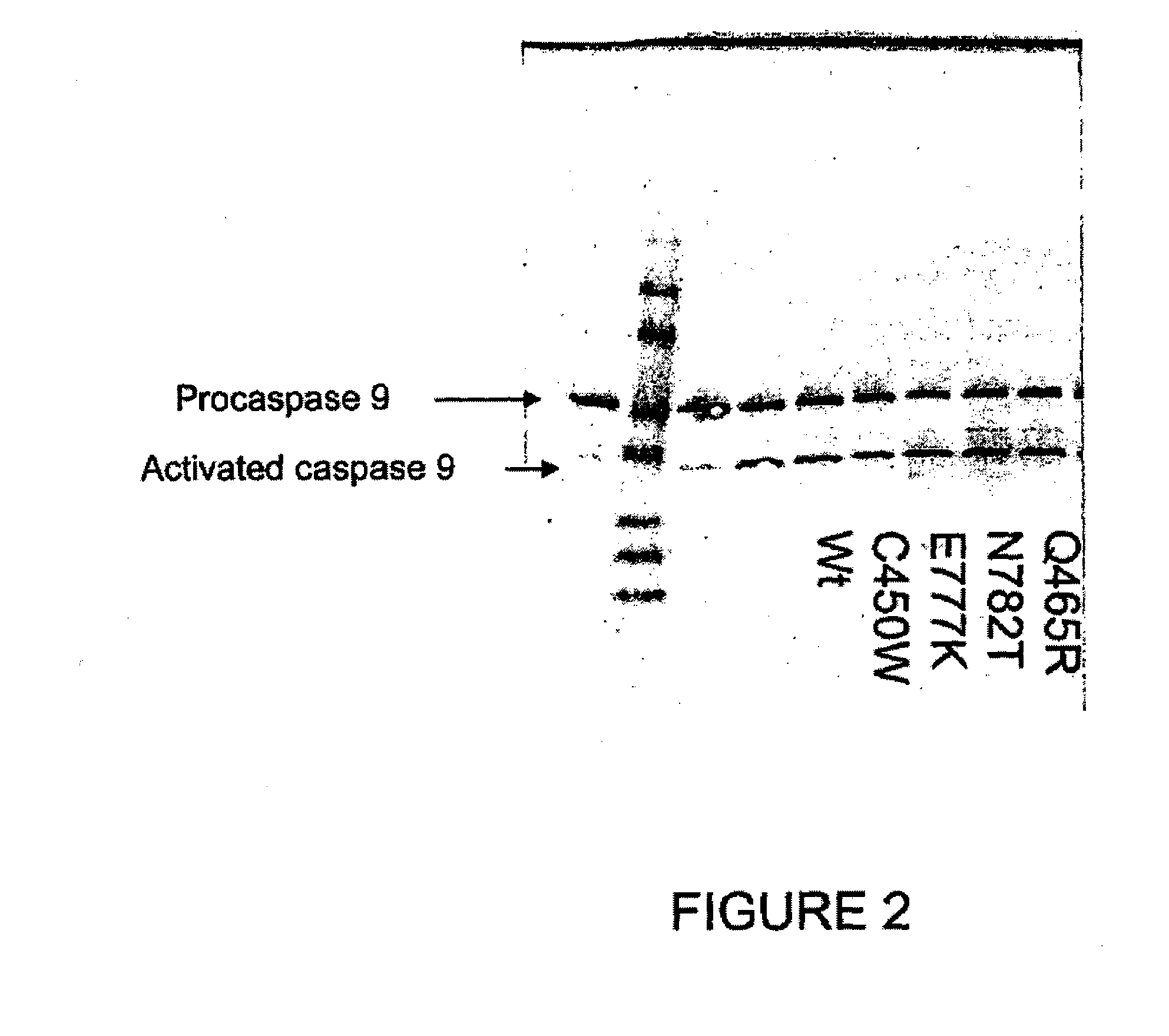
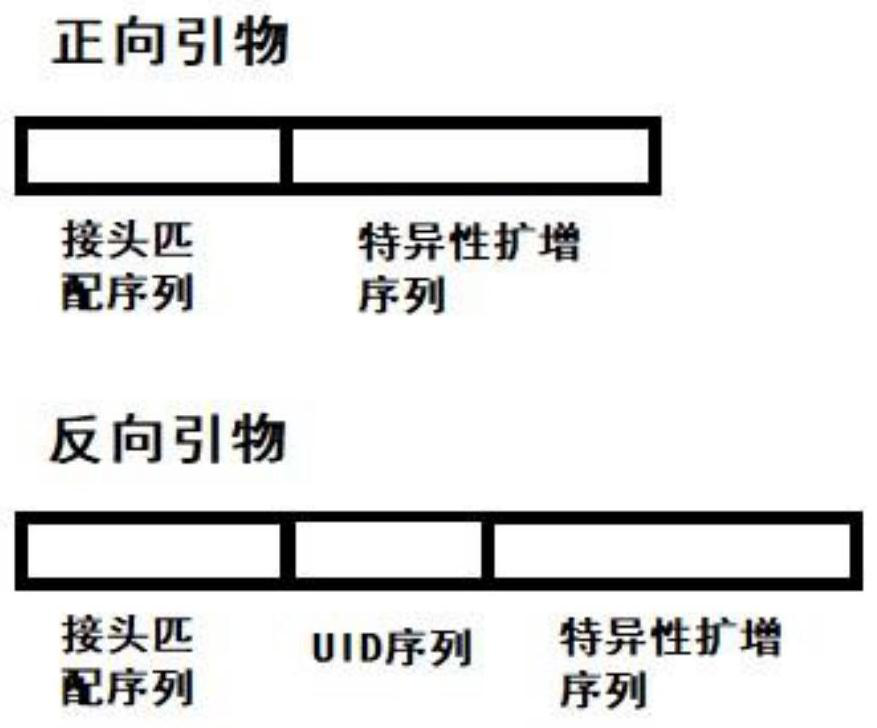
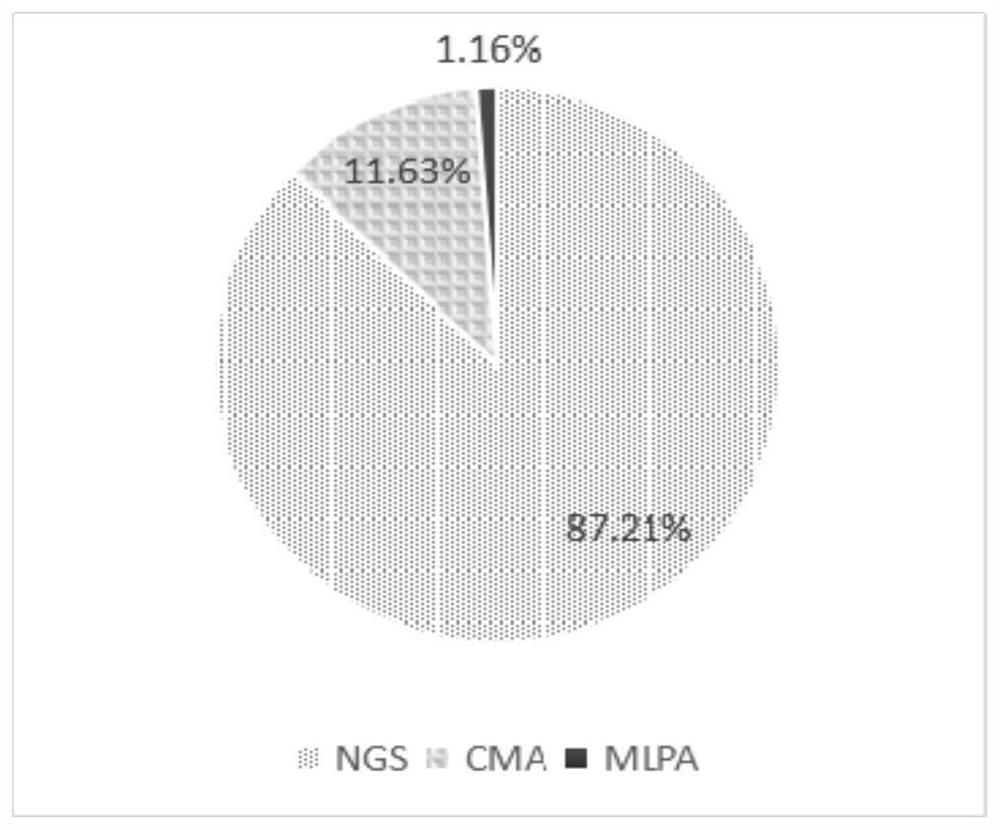
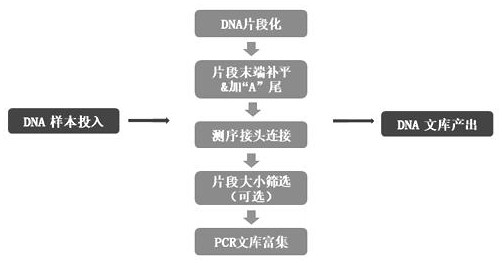
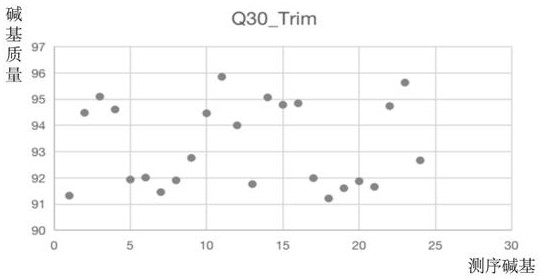
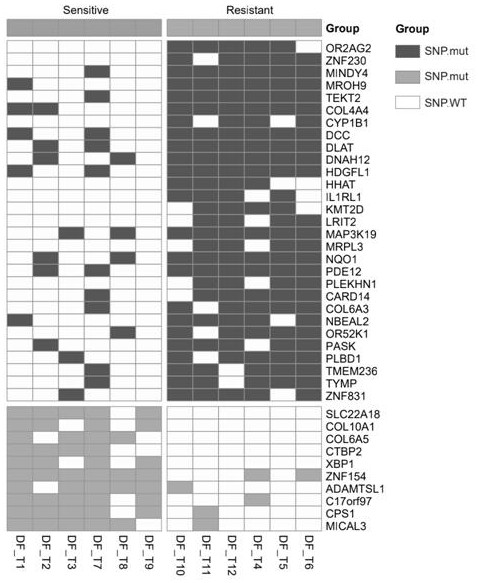
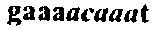Patents
Literature
35 results about "Germline mutation" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
A germline mutation, or germinal mutation, is any detectable variation within germ cells (cells that, when fully developed, become sperm and ovum). Mutations in these cells are the only mutations that can be passed on to offspring, when either a mutated sperm or oocyte come together to form a zygote. After this fertilization event occurs, germ cells divide rapidly to produce all of the cells in the body, causing this mutation to be present in every somatic and germline cell in the offspring; this is also known as a constitutional mutation. Germline mutation is distinct from somatic mutation.
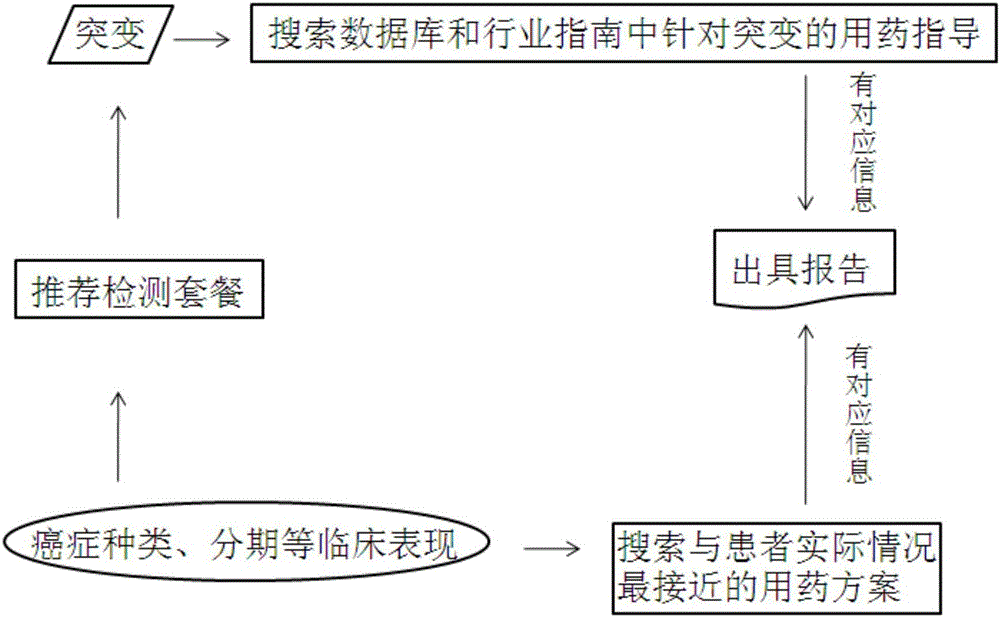
Intelligent tumor medication instruction system
InactiveCN106326652AConvenient guidanceQuick guideSpecial data processing applicationsICT adaptationGermline mutationGenes mutation
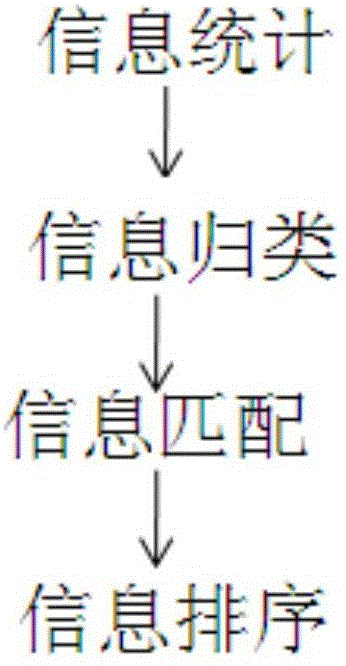
The invention relates to an intelligent tumor medication instruction system. The intelligent tumor medication instruction system is characterized by comprising a patient information module, a gene mutation detection module, a medication instruction database and a detection result reporting module, wherein the patient information module is used for recording the basic information of a patient; the gene mutation detection module is used for detecting the gene mutation information of the patient, such as a gene name, a mutation site, mutation conditions, mutation frequency and germline mutation and / or somatic cell mutation; the medication instruction database is used for performing medication instruction and query according to the clinical information of the patient, automatically matching and sequencing the clinical information and the medication instruction and query results, and sending an optimum medication instruction scheme suitable for the tumor patient to the detection result reporting module; a doctor can instruct the medication on the tumor patient according to the medication scheme displayed by the detection result reporting module. The intelligent tumor medication instruction system is widely applied to the diagnosis and medication instruction process of the tumor patient.
Owner:BEIJING CAPITALBIO MEDLAB CO LTD +1
Detecting somatic single nucleotide variants from cell-free nucleic acid with application to minimal residual disease monitoring
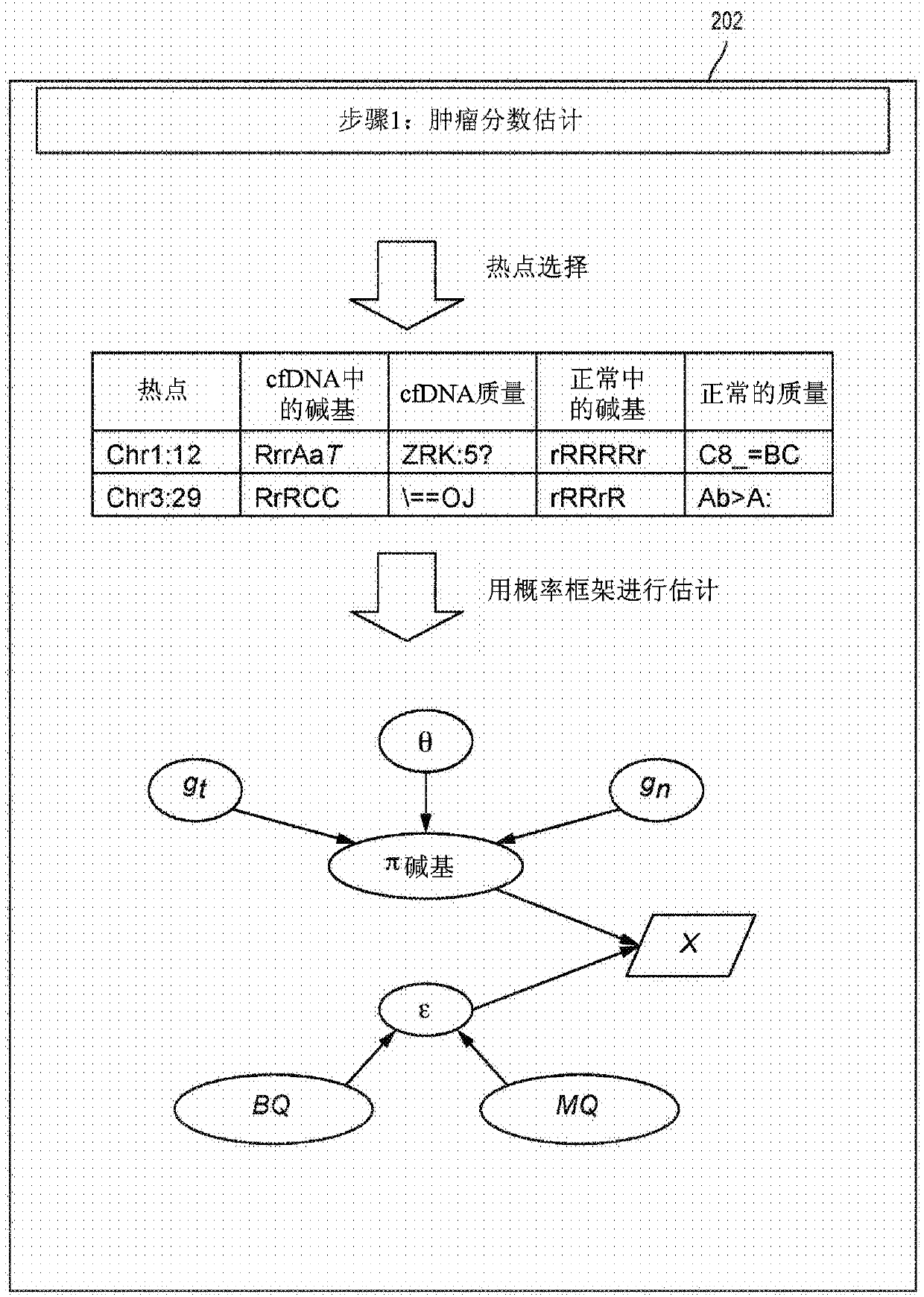
The present disclosure provides a probabilistic model for accurate and sensitive somatic single nucleotide variant (SNV) detection in cell-free nucleic acid samples comprising a set of sequence data.A joint genotype may be determined for each locus in the set of sequence data, and germline mutations may be intrinsically removed. A set of filtrations can be applied to eliminate low quality somaticvariant calls. Further, a global tumor cell-free deoxyribonucleic acid (cfDNA) fraction and overlapping read mates can be considered, thereby enabling accurate SNV detection and variant allele frequency estimation from samples with low tumor cfDNA fraction. A sensitive early detection of minimal residual disease (MRD) is designed by using the probabilistic model and the machine learning model fordistinguishing true variants from sequencing errors.
Owner:RGT UNIV OF CALIFORNIA
Determining a predisposition to cancer
InactiveUS7319007B2Increased susceptibilityMicrobiological testing/measurementAnimal husbandryGermline mutationOncology
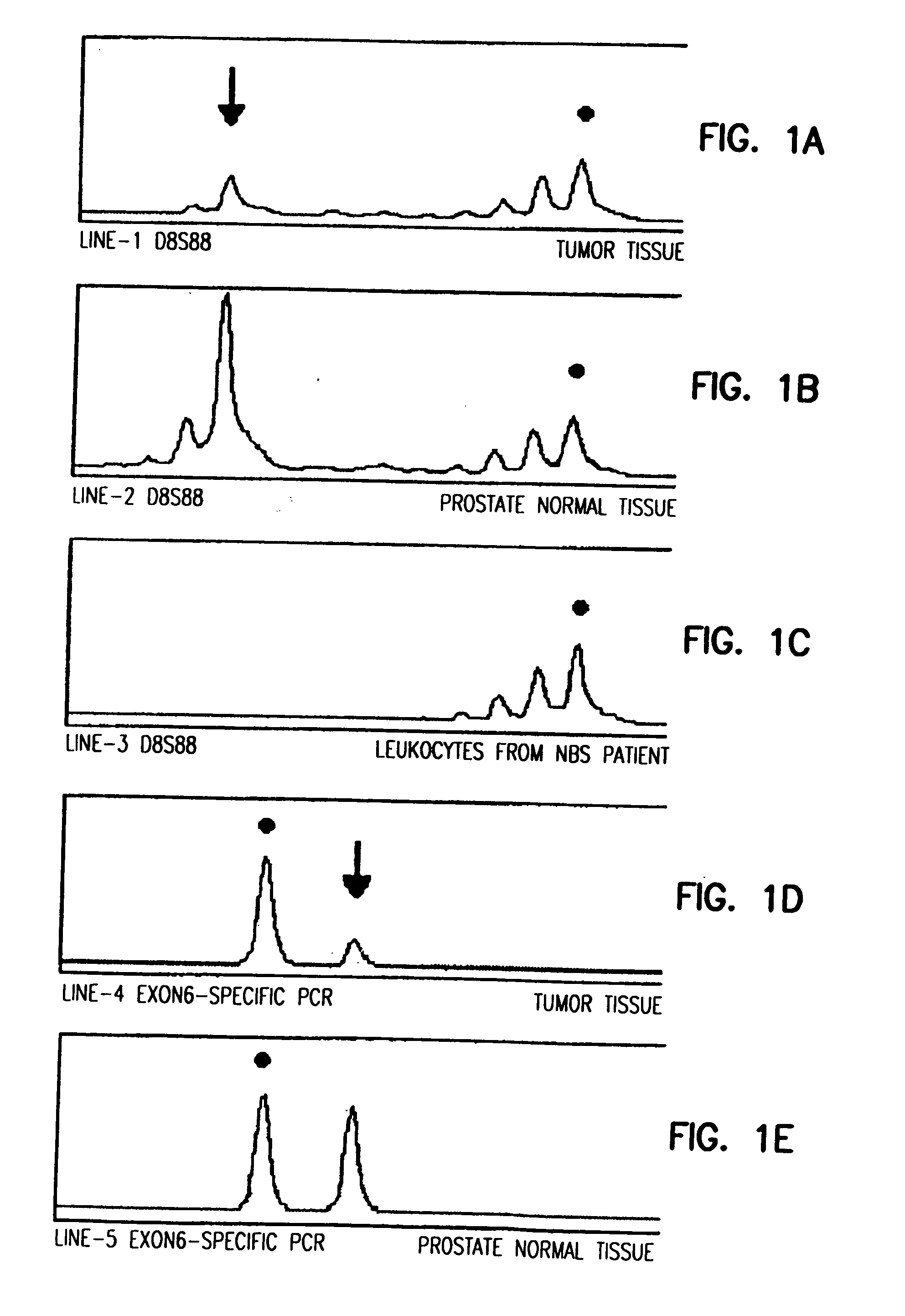
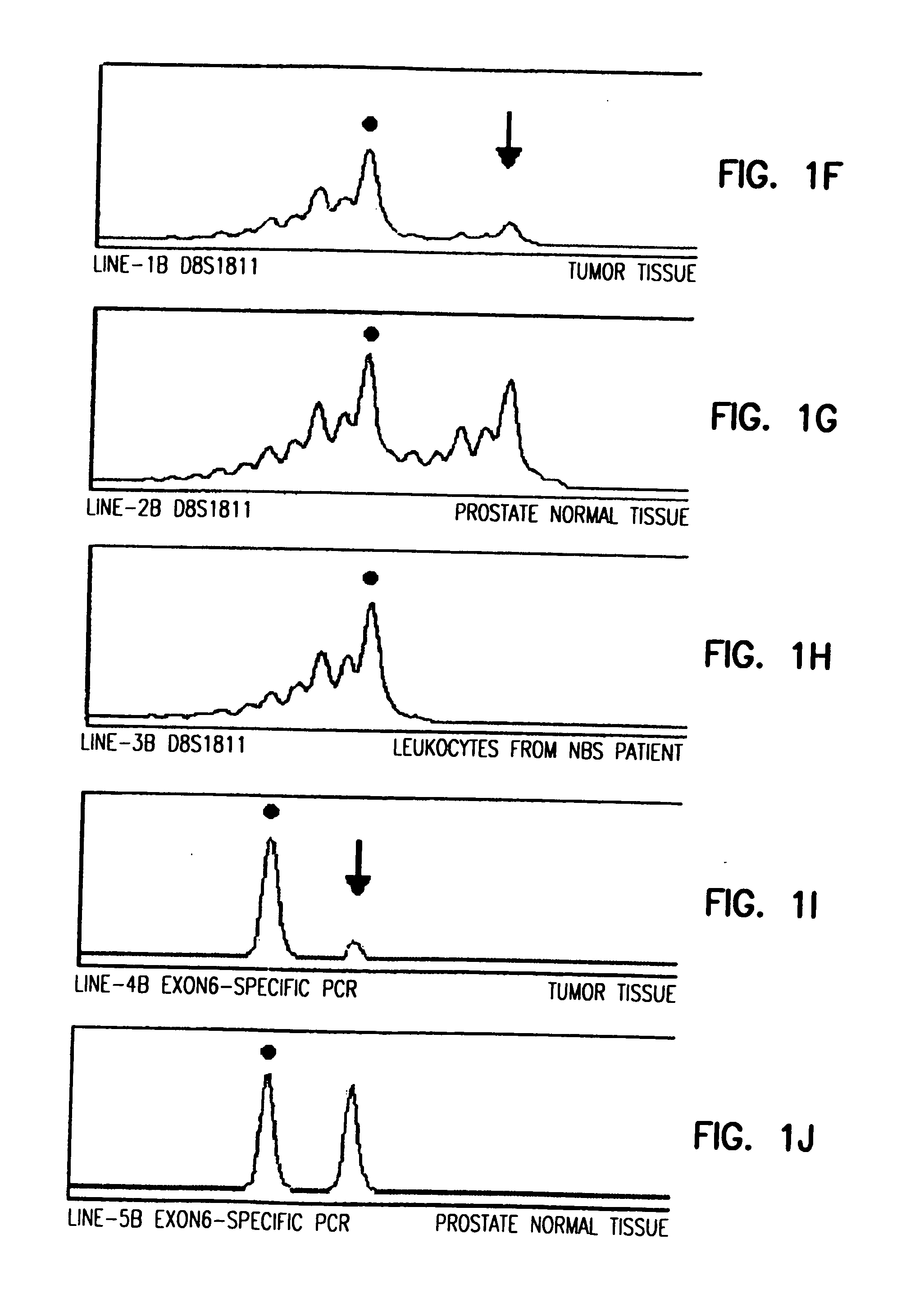
The present invention relates to methods and kits for determining a predisposition for developing cancer, e.g., prostate and / or breast cancer, due to a germline mutation of a NBS1 gene. The present invention also relates to surveillance protocols for developing cancer, e.g., prostate and / or breast cancer, due to germline mutation of a NBS1 gene.
Owner:POMERANIAN ACAD OF MEDICINE
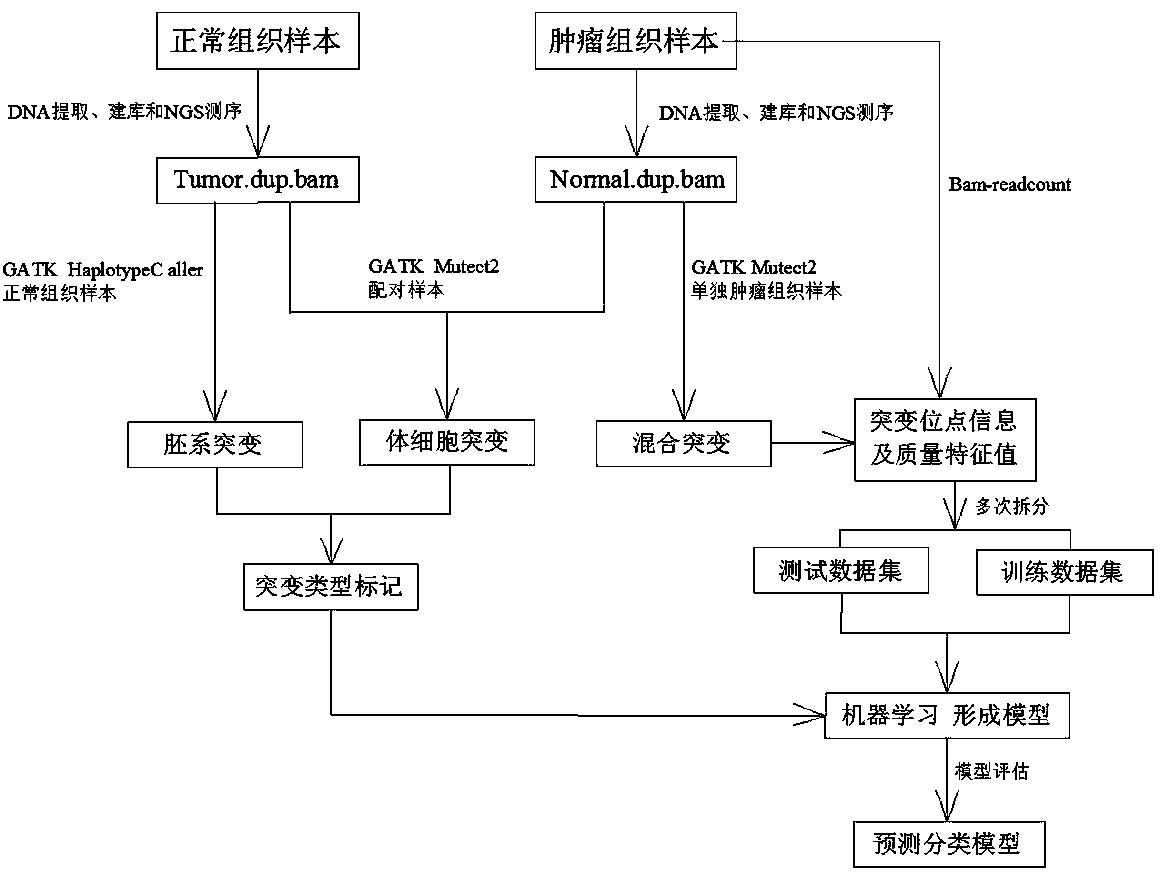
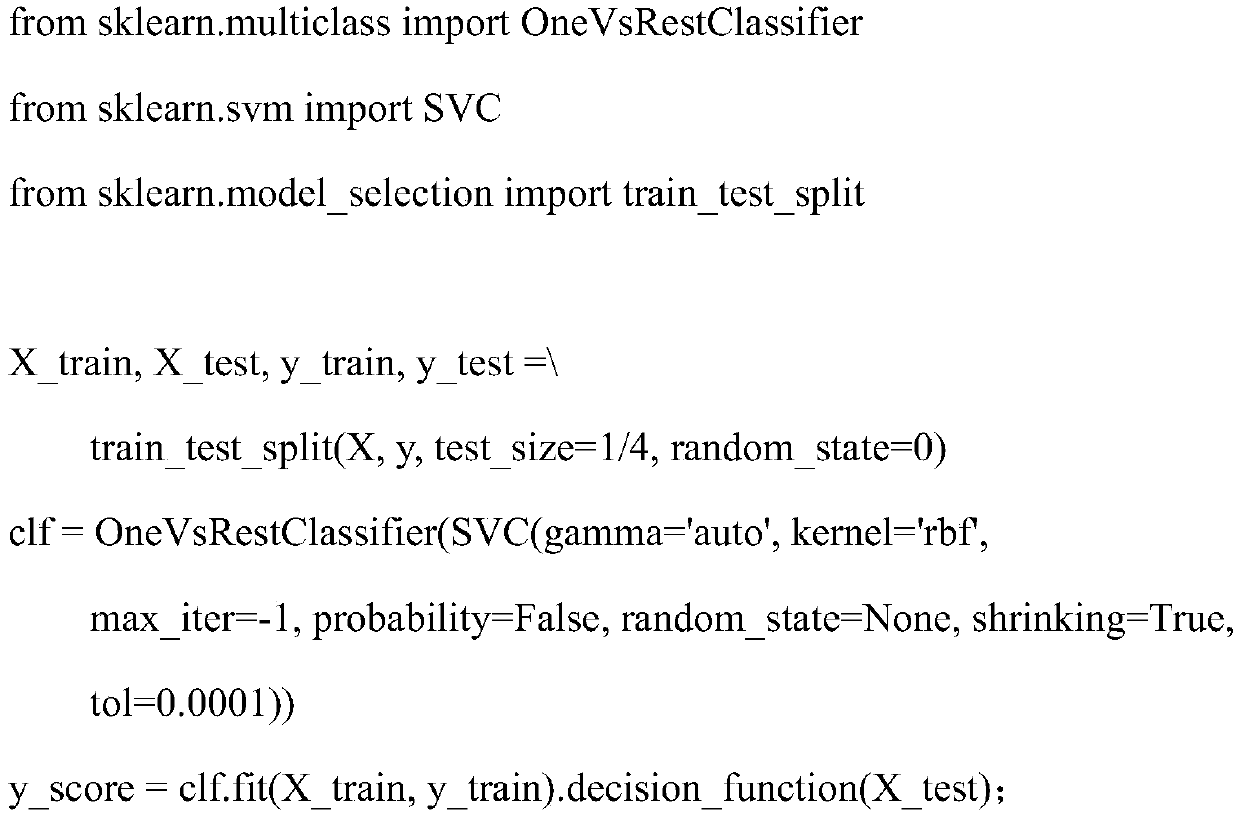
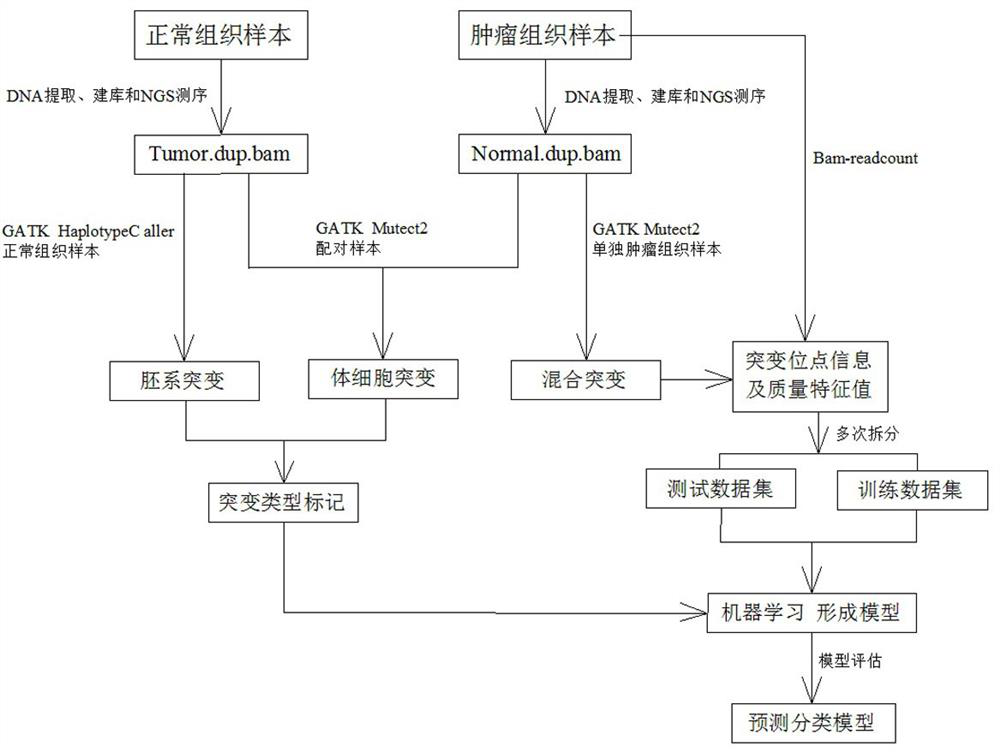
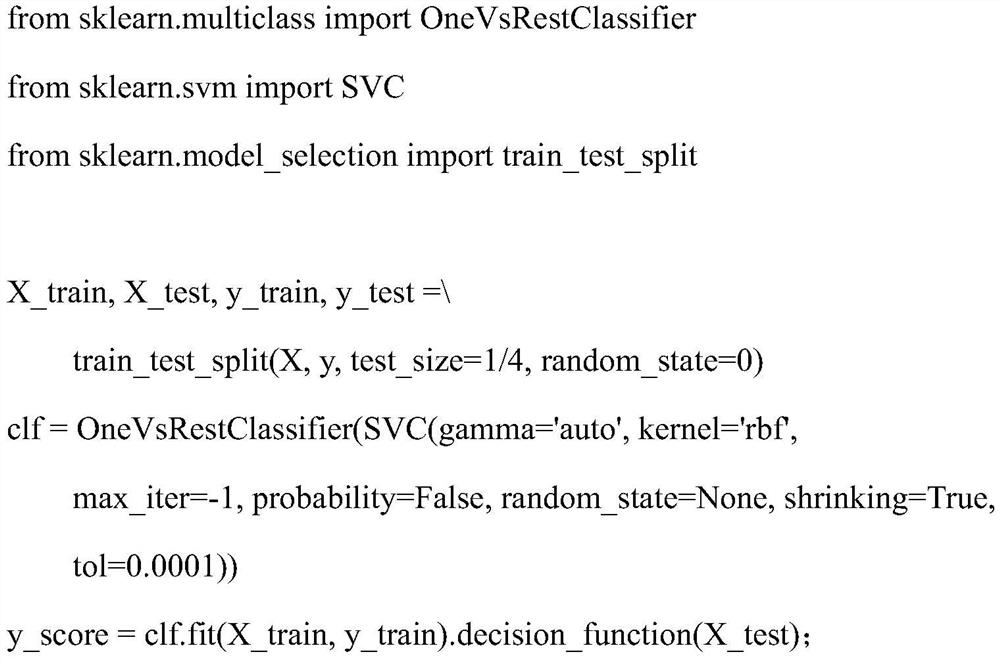
Method for distinguishing gene mutation type from individual tumor sample based on second-generation sequencing
ActiveCN110846411ALow detection costImprove detection efficiencyMicrobiological testing/measurementProteomicsGermline mutationSomatic cell
The invention relates to a method for distinguishing gene mutation types an individual tumor sample based on second-generation sequencing. A tumor tissue sample and a normal tissue sample are used forlibrary construction and NGS sequencing respectively, strand bias and different types of base frequencies of mutation sites stored in an intermediate file BAM for biological information analysis of the tumor tissue sample are analyzed, the quality of base comparison and the frequency of noise are used as the training characteristics of machine learning, meanwhile, type information of corresponding mutation sites of the normal tissue sample is paired to serve as a prediction mutation type, a classification prediction model is constructed to distinguish somatic mutation from germline mutation,the model is used to distinguish somatic mutation from germline mutation, the detection efficiency is high, the specificity is high, and after the model is established, the individual tumor sample canbe used for NGS sequencing and mutation detection, the detection cost of a normal or cancer sample can be well saved, and meanwhile, the problem that normal tissues of tumor patients with specific types are difficult to obtain can be solved.
Owner:上海仁东医学检验所有限公司 +1
Depression gene
InactiveUS7410944B1Avoid depressionImprove disease symptomsPeptide/protein ingredientsDepsipeptidesGermline mutationGenetics human
Owner:MYRIAD GENETICS
Determining a predisposition to cancer
The present invention relates to methods and kits for determining a predisposition for developing cancer, e.g., prostate and / or breast cancer, due to a germline mutation of a NBS1 gene. The present invention also relates to surveillance protocols for developing cancer, e.g., prostate and / or breast cancer, due to germline mutation of a NBS1 gene.
Owner:POMERANIAN ACAD OF MEDICINE
Obesity gene and use thereof
InactiveUS20080085516A1Prevent obesityImprove disease symptomsSugar derivativesMicrobiological testing/measurementGermline mutationDiabetes mellitus
The present invention relates generally to the field of human genetics. Specifically, the present invention relates to methods and materials used to isolate and detect a human obesity and diabetes predisposing gene, specifically the TBC1D1 gene, some mutant alleles of which cause susceptibility to obesity and / or diabetes. More specifically, the invention relates to germline mutations in the TBC1D1 gene and their use in the diagnosis of predisposition to obesity and diabetes. Finally, the invention relates to the screening of the TBC1D1 gene for mutations / alterations, which are useful for diagnosing the predisposition to obesity.
Owner:UNIV OF UTAH RES FOUND
Diagnostic test kit for determining a predisposition for breast and ovarian cancer, materials and methods for such determination
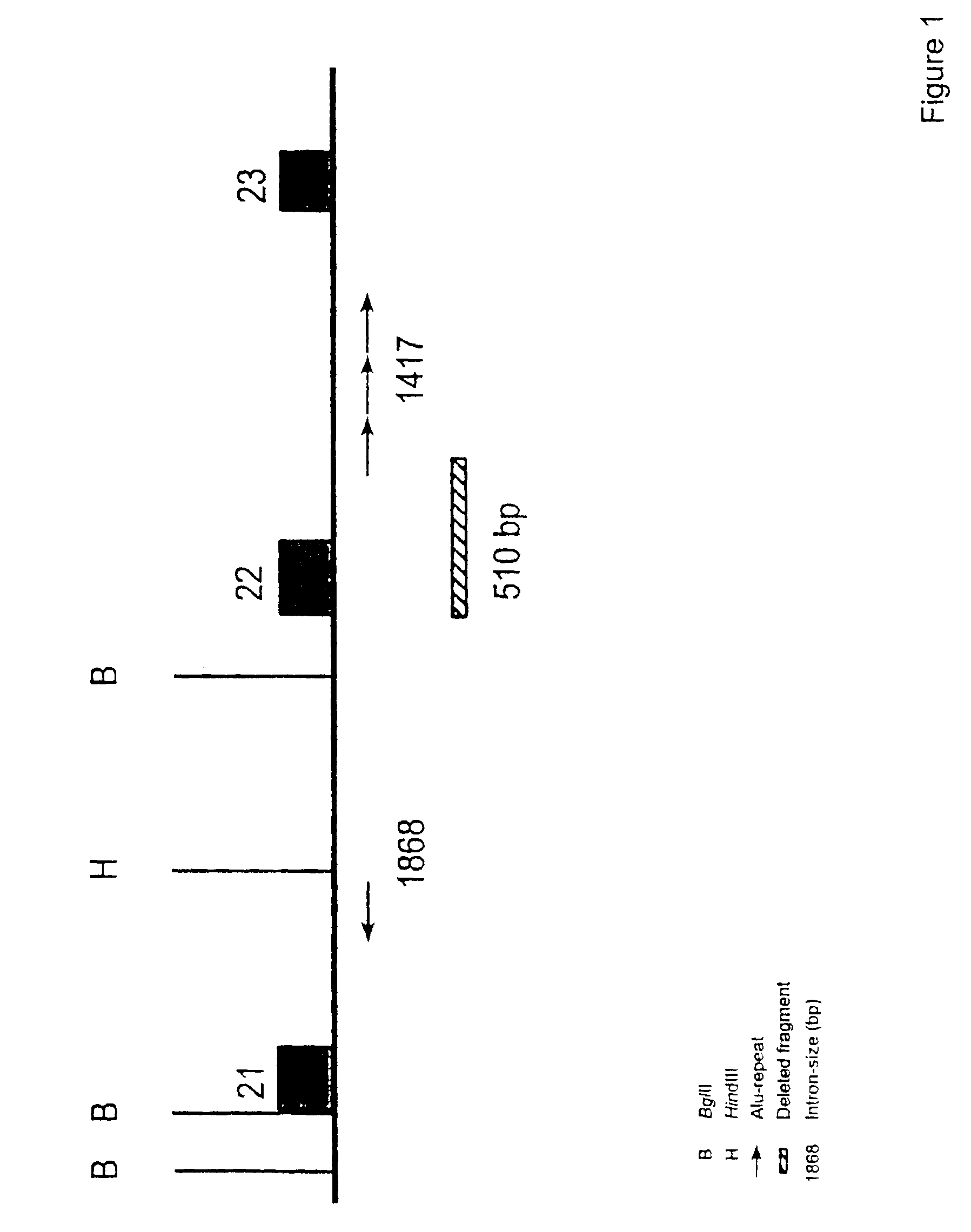
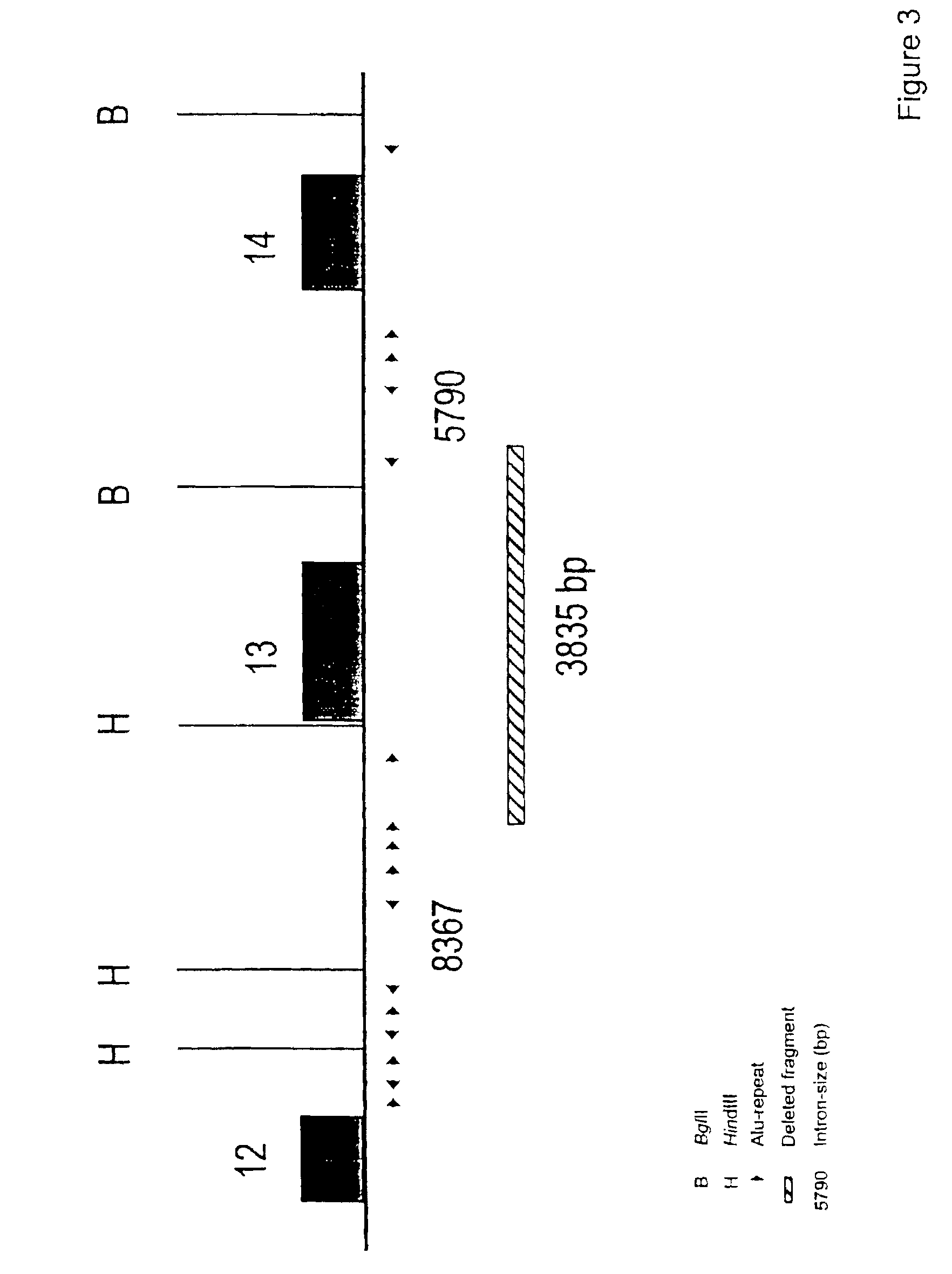
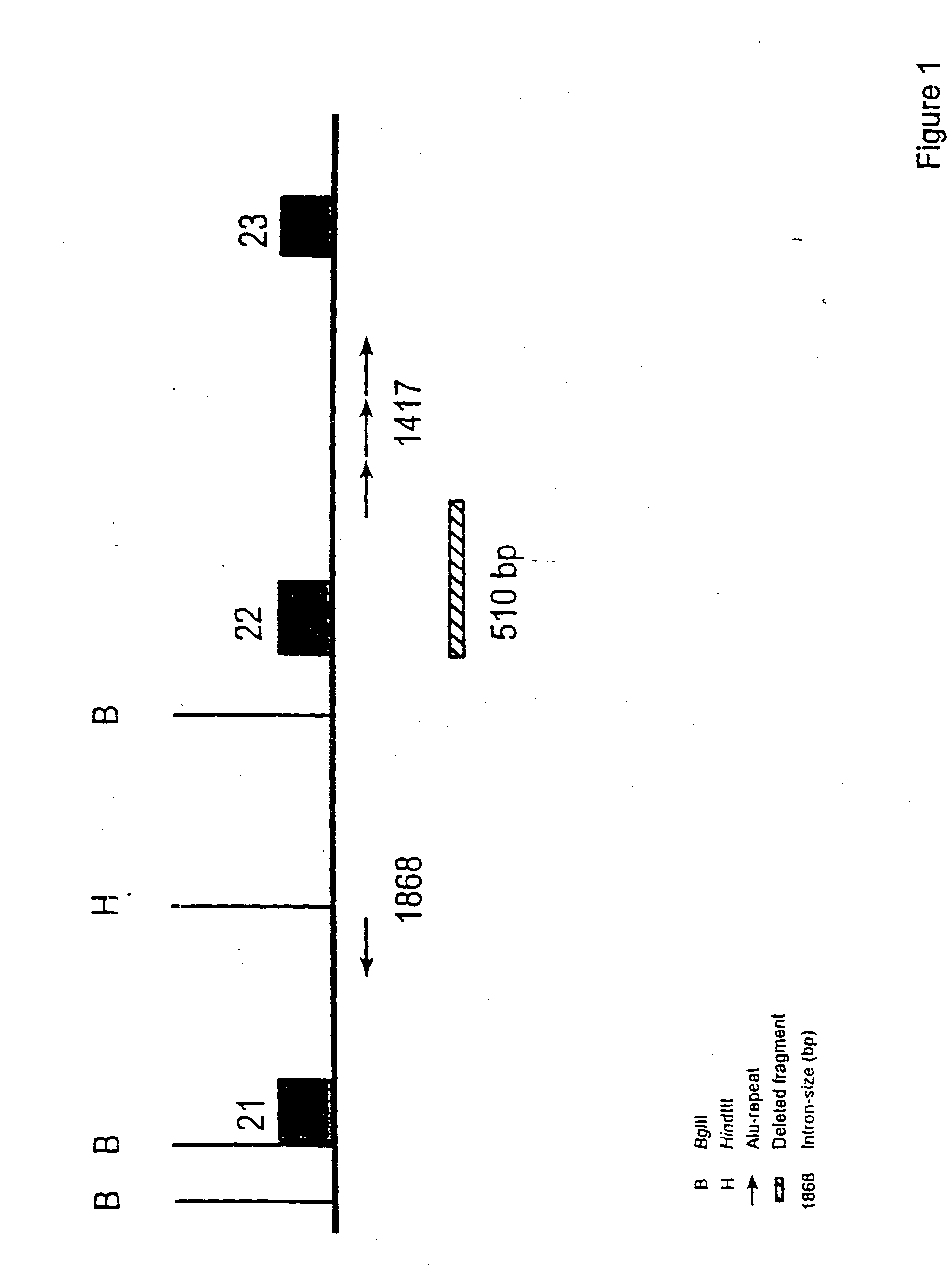
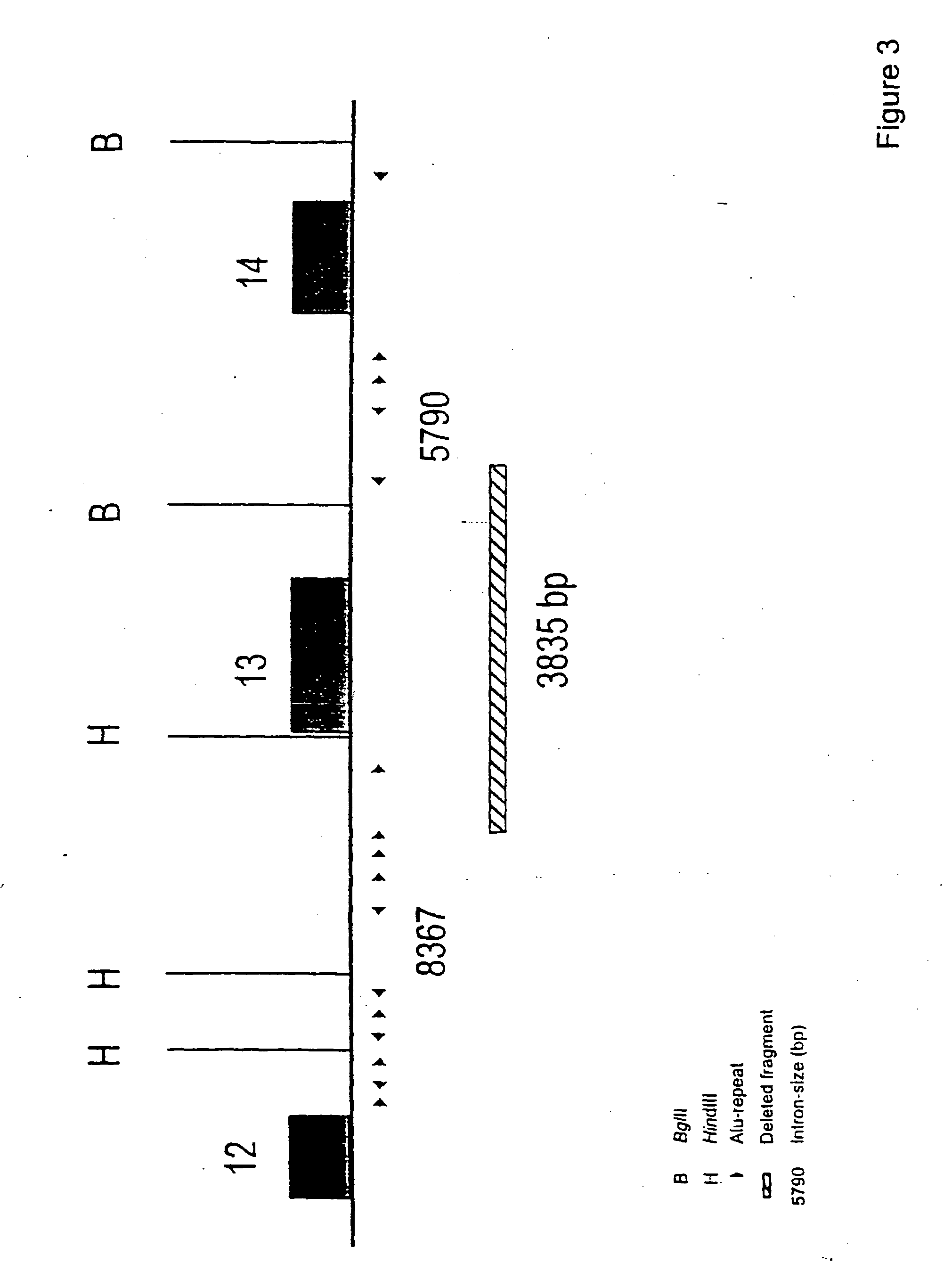
The present invention relates generally to the field of human genetics, and more specifically to the detection of a specific type of germline mutations in the BRCA1 gene, which will predispose to breast and ovarian cancer. In addition, the invention relates to the molecular genetic mechanism that may have mediated the genesis of these mutations, in particular the role of Alu repetitive DNA elements present in the intronic regions of BRCA1. The invention further relates to somatic mutations of this type in the BRCA1 gene in human breast and ovarian cancer, and their use in the diagnosis and prognosis of human breast and ovarian cancer. The invention more particularly relates to the screening of this type of BRCA1 mutations in human genomic DNA, which are useful for the diagnosis of inherited predisposition to breast and ovarian cancer.
Owner:LEIDEN UNIVERSITY
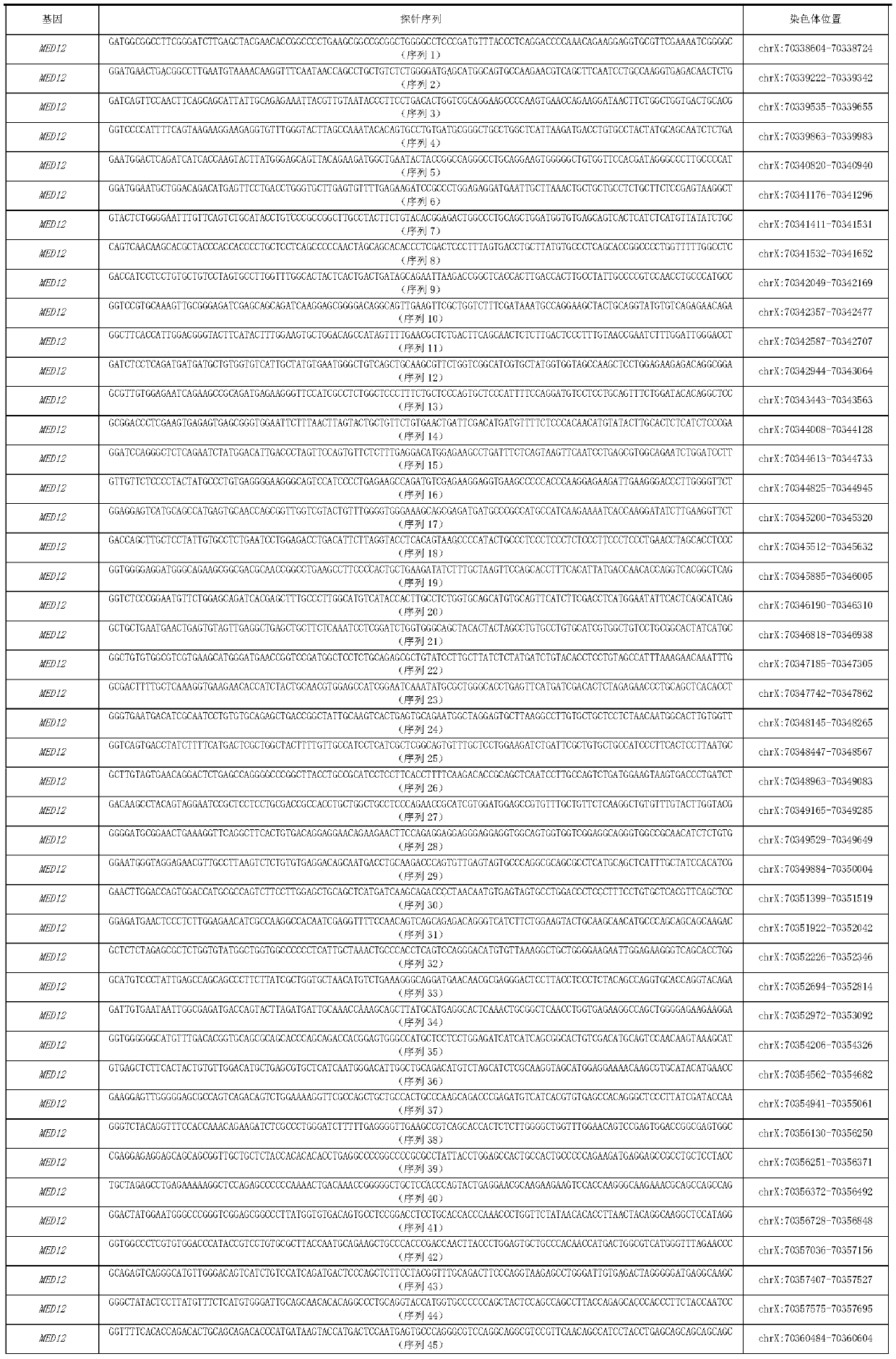
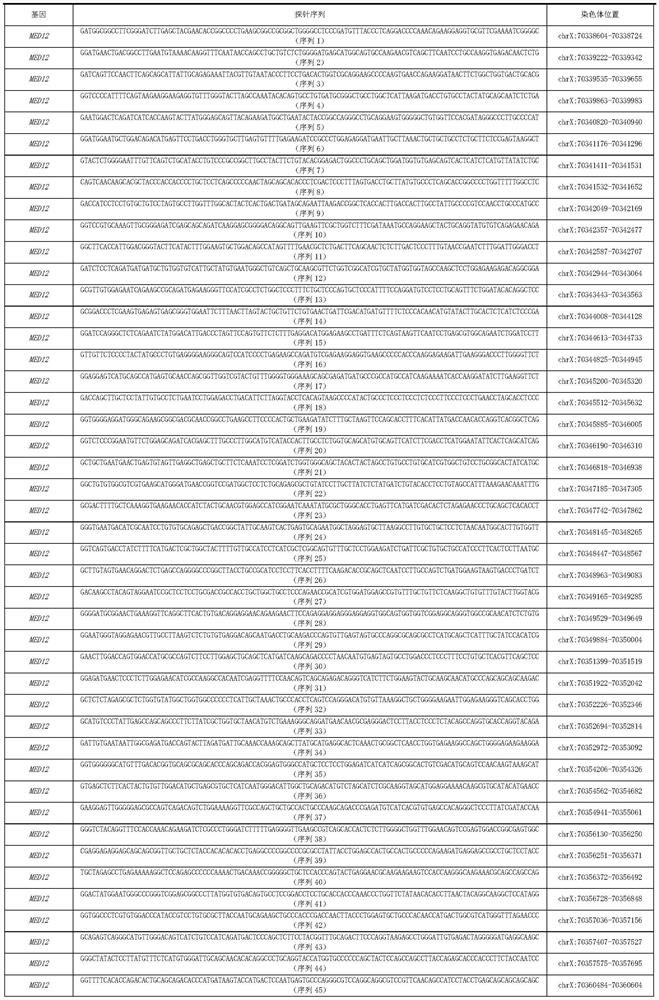
MED12 gene mutation detecting kit and application thereof
ActiveCN110229897AThe detection method is simpleSensitive detection methodMicrobiological testing/measurementDNA/RNA fragmentationMRD NegativeHematological Tumor
The present invention discloses a MED12 gene mutation detecting kit and an application thereof. The kit comprises probe sets for detecting MED12 gene mutations and the probe sets consist of a probe 1to a probe 48. The kit can be used for detecting MED12 somatic mutations and germline mutations at a tumor genome level, can also be used for assistant diagnosis and minimal residual disease monitoring and prognosis evaluation, etc. of patients with hematological tumors, especially patients with acute myeloid leukemia. Besides, the kit is simple, sensitive and accurate in detection method and plays an important role in the field of medical detection
Owner:PEOPLES HOSPITAL PEKING UNIV
Depression gene
InactiveUS7052853B1Avoid depressionImprove disease symptomsPeptide preparation methodsDepsipeptidesGermline mutationGenetics human
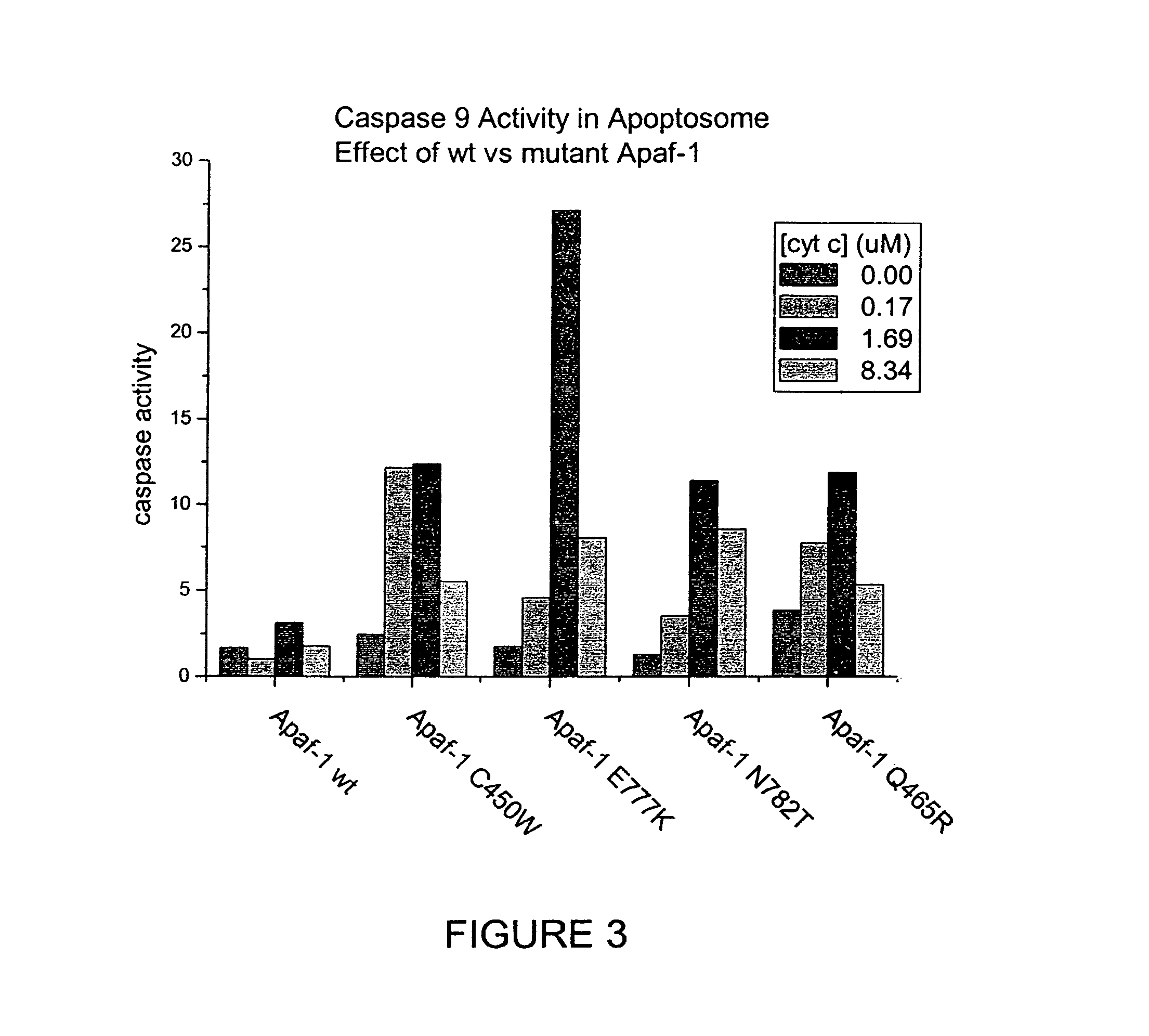
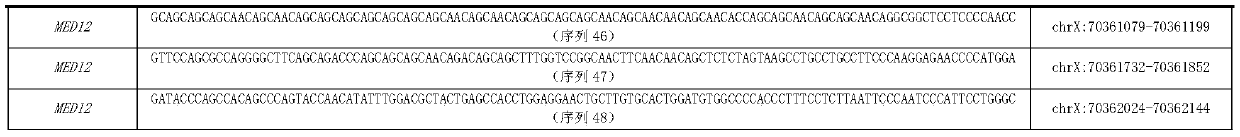
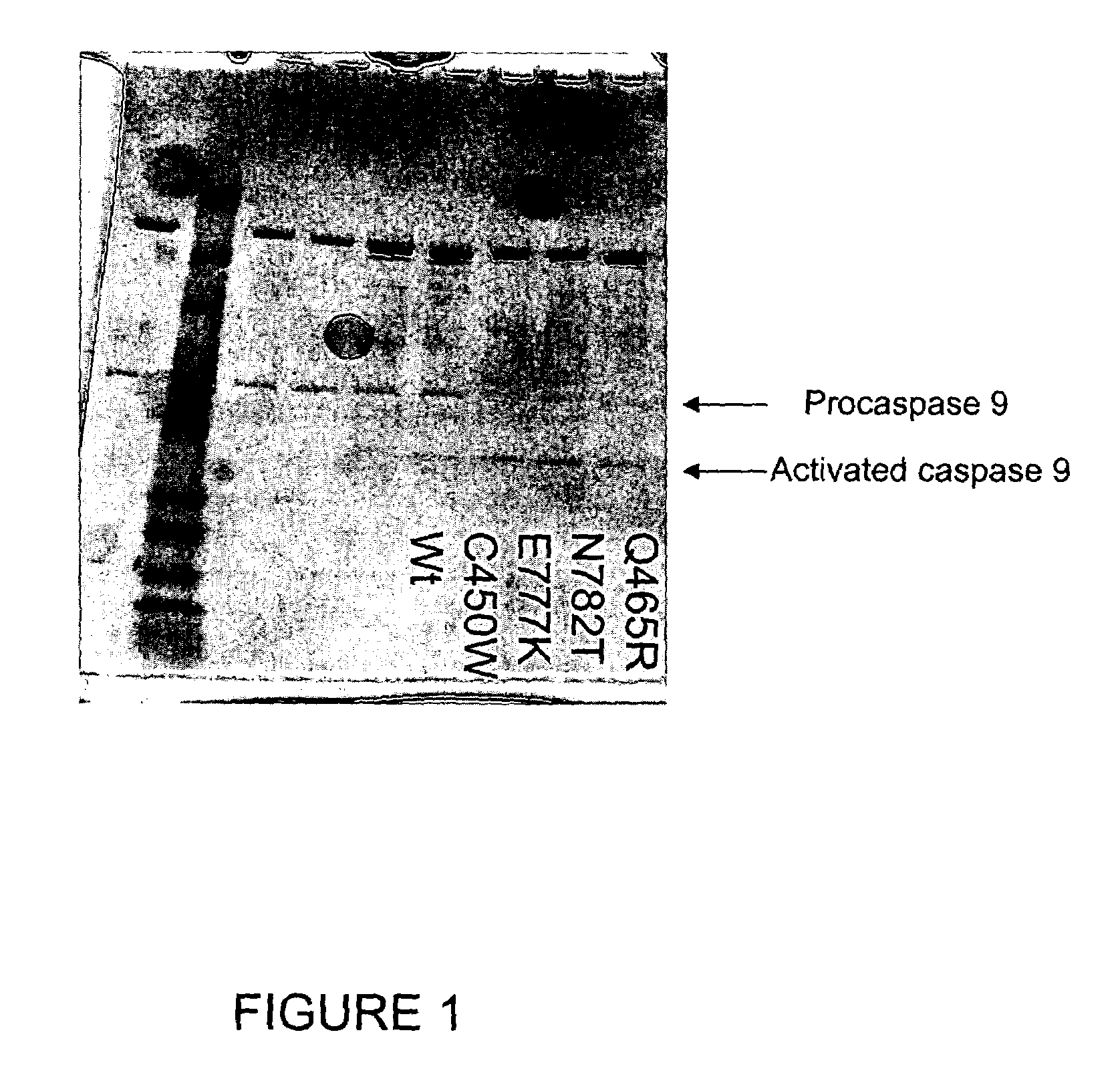
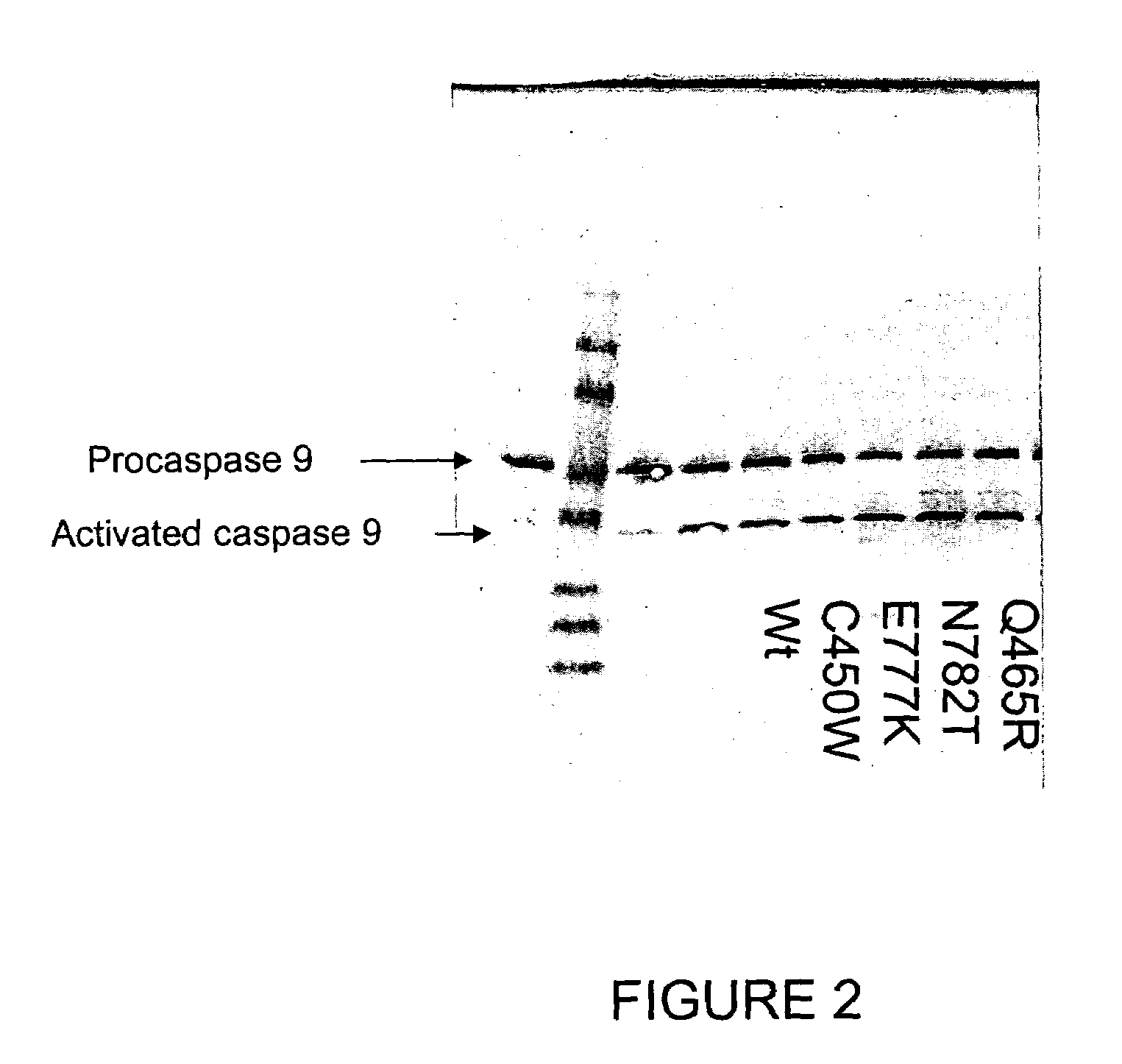
The present invention relates generally to the field of human genetics. Specifically, the present invention relates to methods and materials used to isolate and detect a human depression predisposing gene, specifically the apoptotic protease activating factor 1 (APAF1) gene, some mutant alleles of which cause susceptibility to depression. More specifically, the invention relates to germline mutations in the APAF1 gene and their use in the diagnosis of predisposition to depression. The invention also relates to the prophylaxis and / or therapy of depression associated with a mutation in the APAF1 gene. The invention further relates to the screening of drugs for depression therapy. Finally, the invention relates to the screening of the APAF1 gene for mutations / alterations, which are useful for diagnosing the predisposition to depression.
Owner:MYRIAD GENETICS
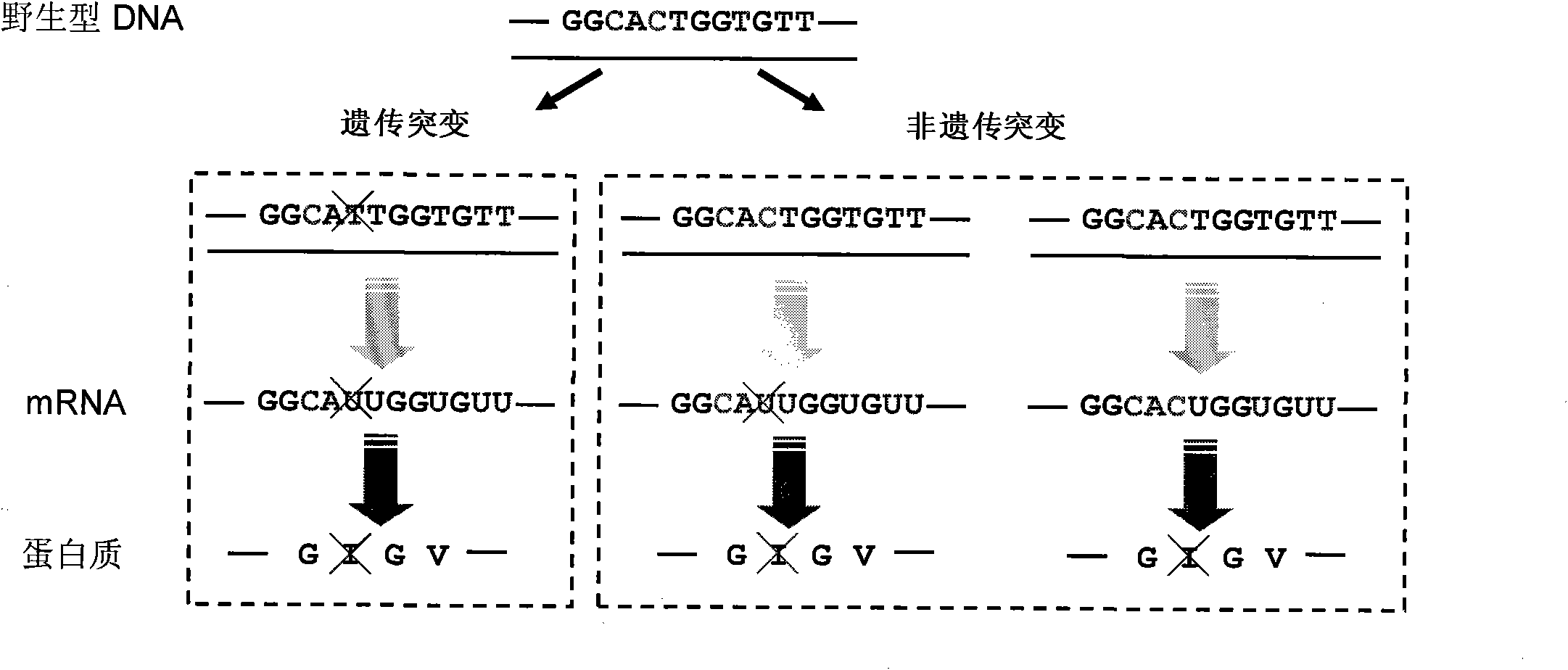
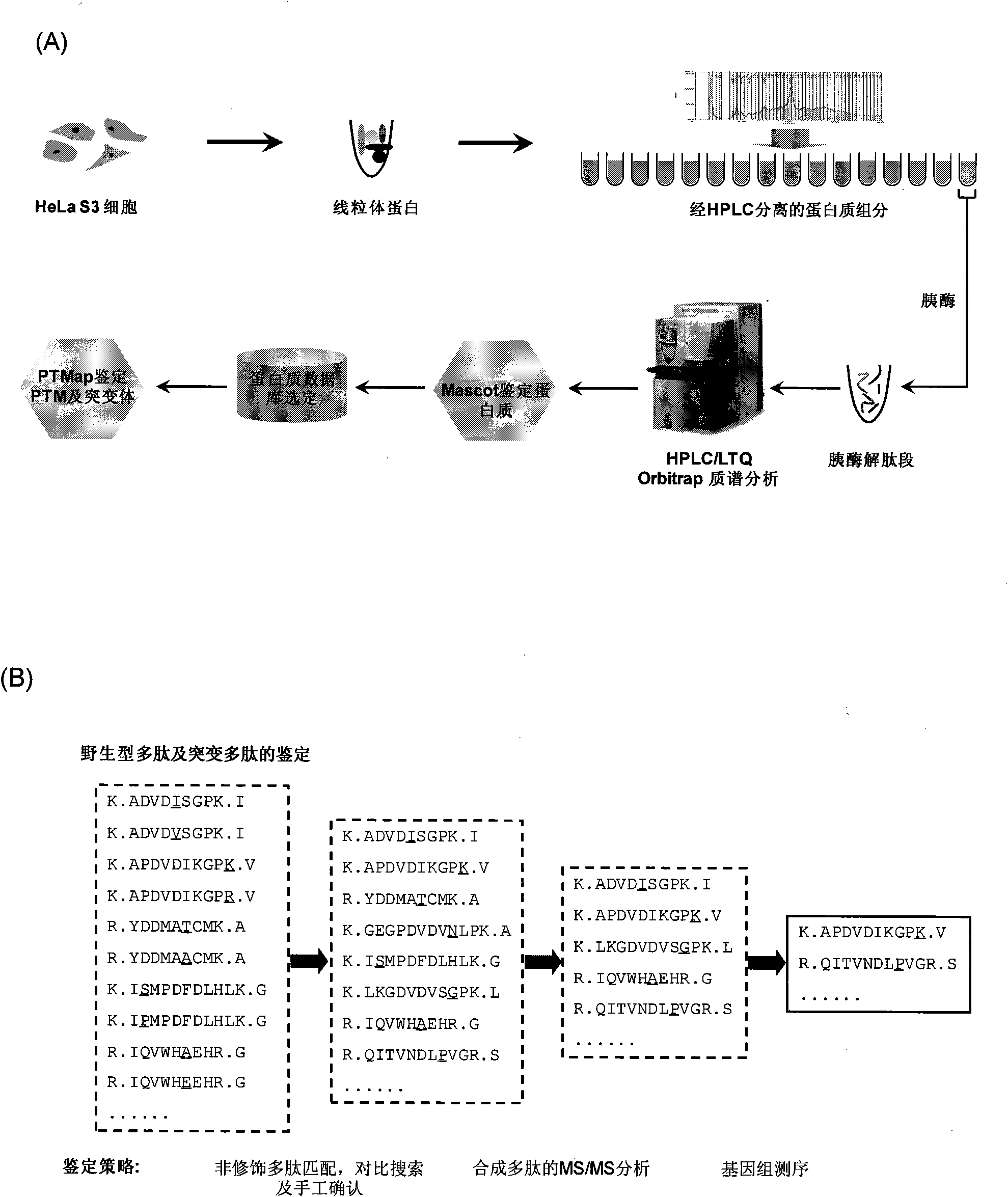
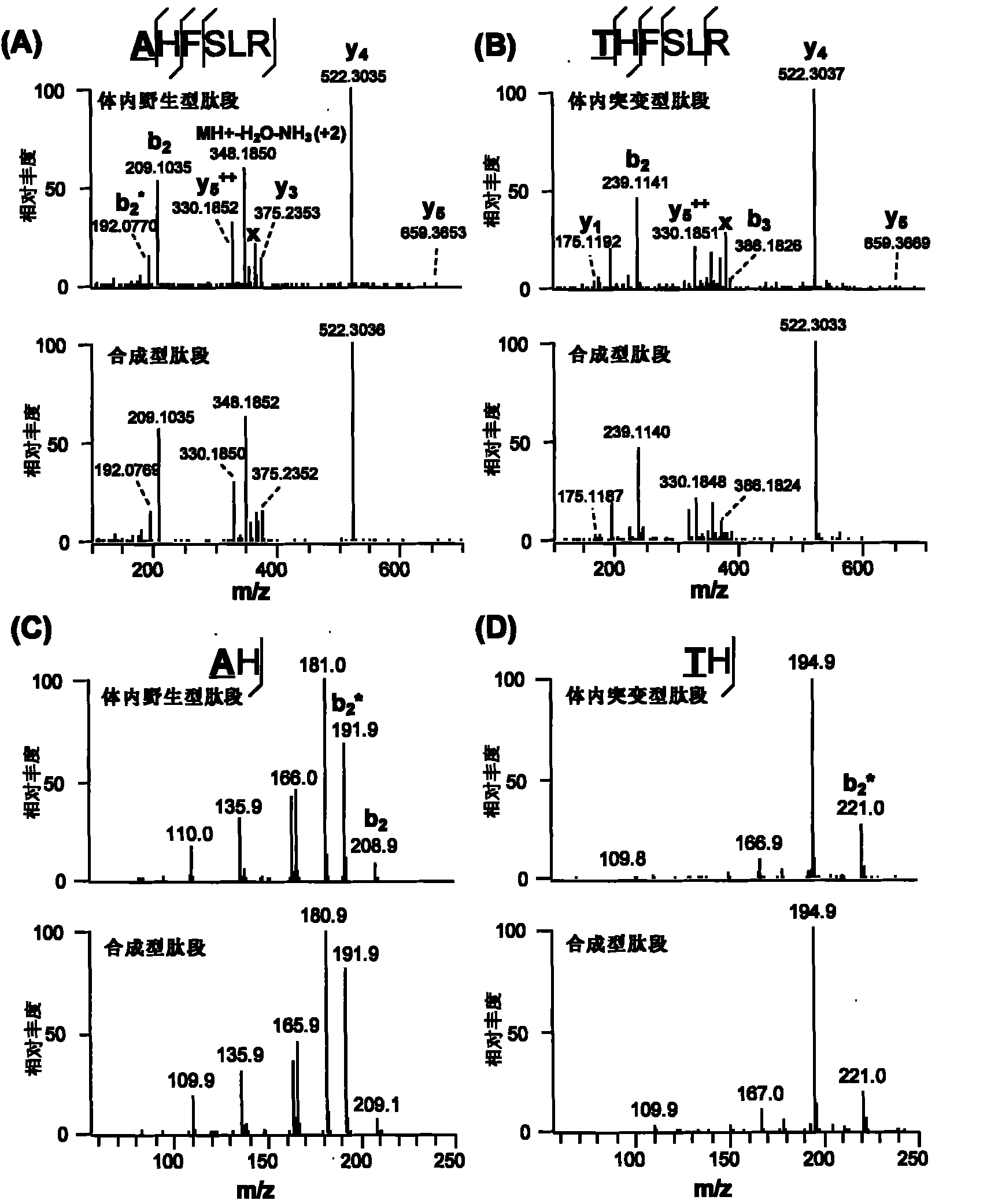
Identification method of highly stoichiometric non-germline mutation in eucaryotic cell
ActiveCN102140496AHigh stoichiometric ratioMicrobiological testing/measurementMaterial analysis by electric/magnetic meansGermline mutationEucaryotic cell
The invention relates to an identification method of the high stoichiometric ratio non-germline mutation in the eucaryotic cell and provides an semi-quantitative identification method of the protein synthesis error caused by the single amino acid mutation of the non-germline mutation (NGM), wherein the method is based on the proteomics technology and genome DNA sequencing of the mass spectrometry. The method is particularly as follows: the large-scale proteome analysis, the unrestricted sequence alignment and the identification and gene sequencing of the high resolution second-order mass spectrum and third-order mass spectrum of the synthetic peptide segment are integrated to perform systematic synthesis to the NGM and identify that the four peptide segments of four proteins contain the NGMs. By adopting the method for estimation, the NGM occurrence in the peptide segment is about 0.05%, which is more than 0.3% on the protein level. According to the peptide mass spectrum count ratio of mutant peptide segments to normal peptide segments, the stoichiometric ratio of the NGM is 0.05 to 0.71. By adopting the identification method, the NGM is proved to have high occurrence and stoichiometric ratio while the NGM is predicted to be used as a possible mechanism to join the basic biological and physiological processes.
Owner:JINGJIE PTM BIOLAB HANGZHOU CO LTD
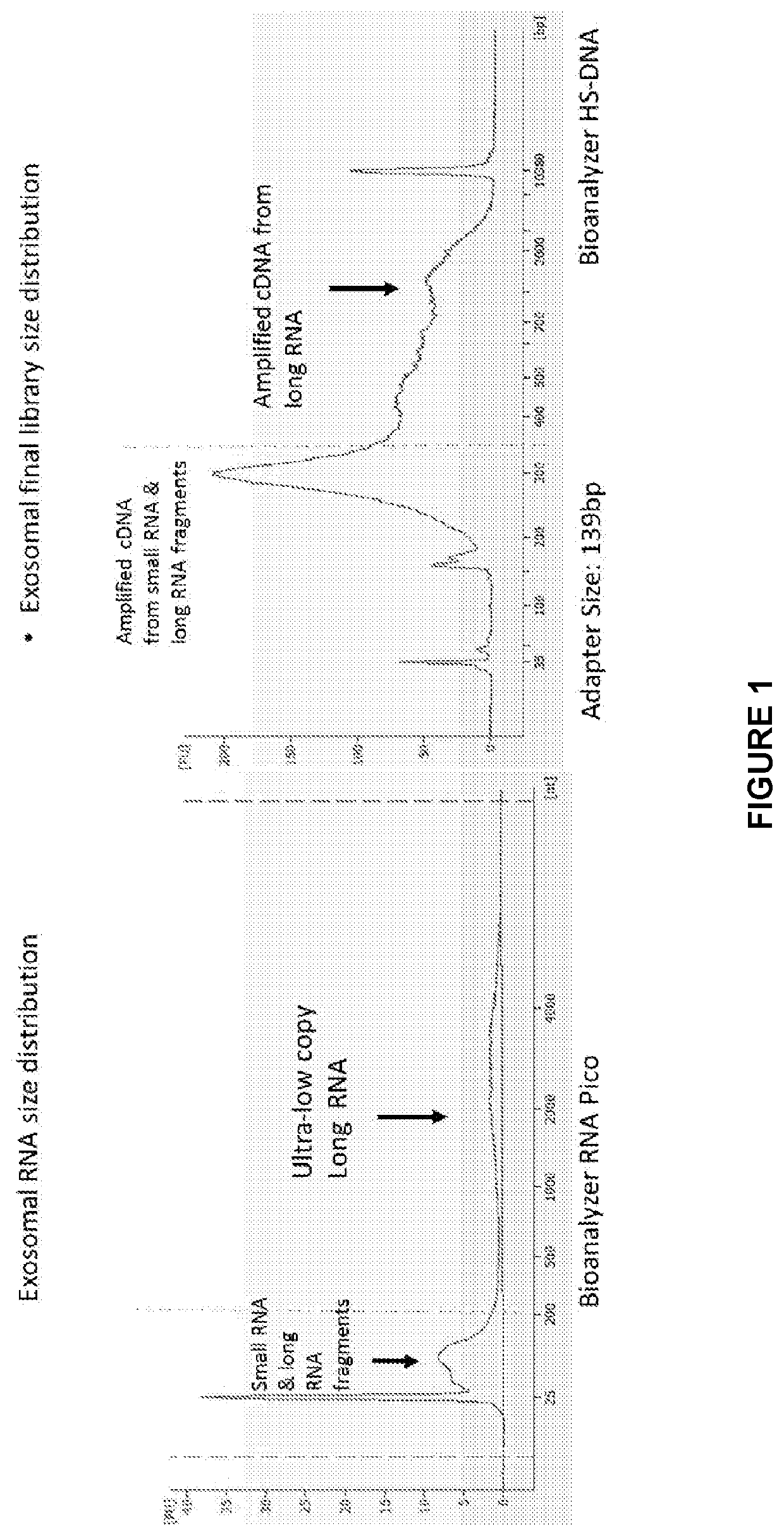
Sequencing and analysis of exosome associated nucleic acids
ActiveUS20200208213A1Microbiological testing/measurementPreparing sample for investigationGermline mutationNucleotide
The invention provides a series of steps that prepare nucleic acids (RNA and / or DNA) isolated from extracellular vesicles for sequencing. This enables a wide diversity of RNAs and / or DNAs, to be efficiently detected. These can then be used to identify various attributes such as gene expression, alternative splicing, and the detection of both somatic and germline mutations including single nucleotide variants (SNV) and structural variations (insertions / deletions, fusions, inversions).
Owner:EXOSOME DIAGNOSTICS
Diagnostic test kit for determining a predisposition for breast and ovarian cancer, materials and methods for such determination
InactiveUS20050019782A1Sugar derivativesMicrobiological testing/measurementGermline mutationDiagnostic test
The present invention relates generally to the field of human genetics, and more specifically to the detection of a specific type of germline mutations in the BRCA1 gene, which will predispose to breast and ovarian cancer. In addition, the invention relates to the molecular genetic mechanism that may have mediated the genesis of these mutations, in particular the role of Alu repetitive DNA elements present in the intronic regions of BRCA1. The invention further relates to somatic mutations of this type in the BRCA1 gene in human breast and ovarian cancer, and their use in the diagnosis and prognosis of human breast and ovarian cancer. The invention more particularly relates to the screening of this type of BRCA1 mutations in human genomic DNA, which are useful for the diagnosis of inherited predisposition to breast and ovarian cancer.
Owner:LEIDEN UNIVERSITY
Diabetes gene
InactiveUS7374884B2Preventing and treating diabetesImprove disease symptomsSugar derivativesPeptide/protein ingredientsInsulin dependent diabetesDiabetes Therapy
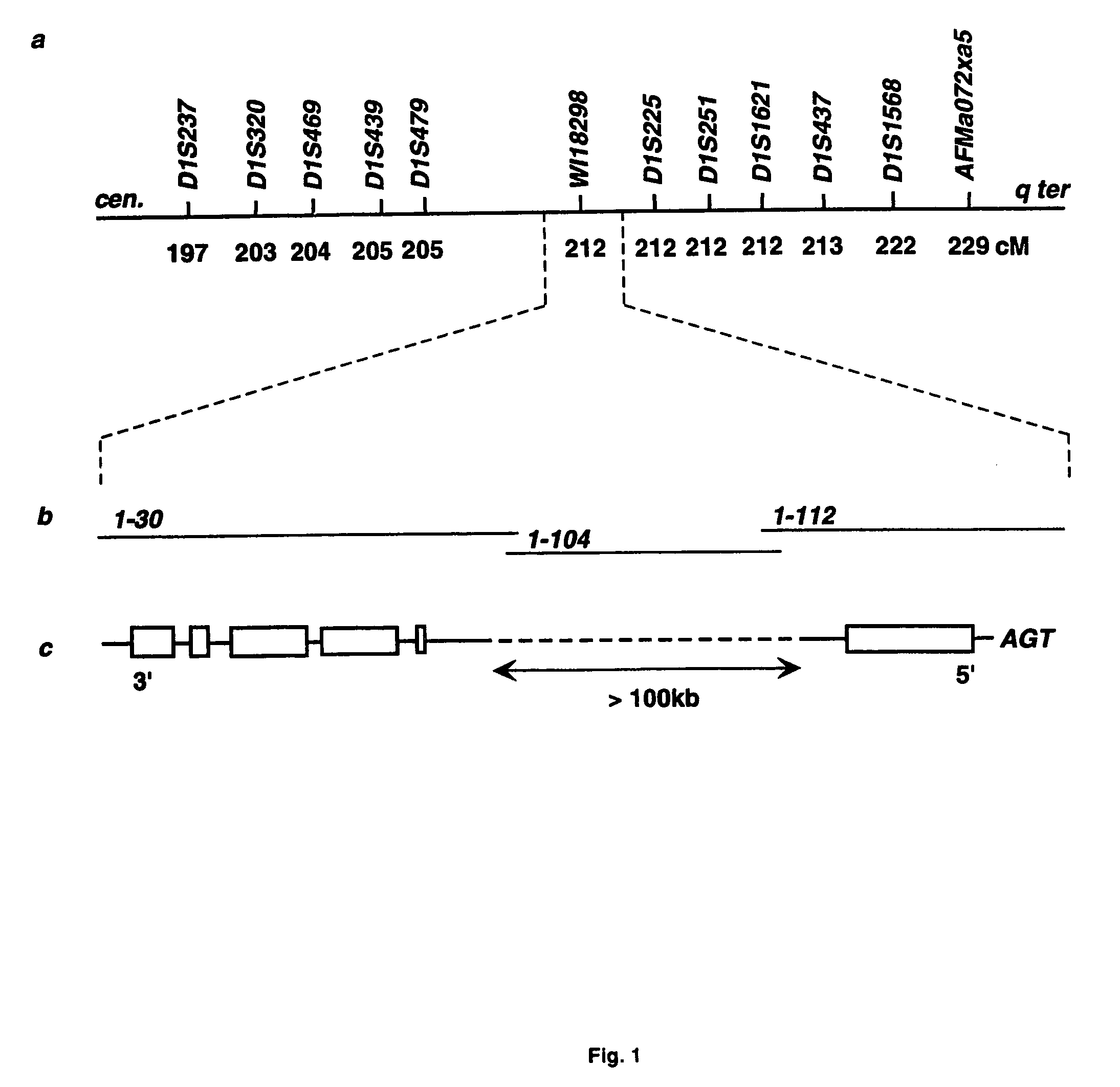
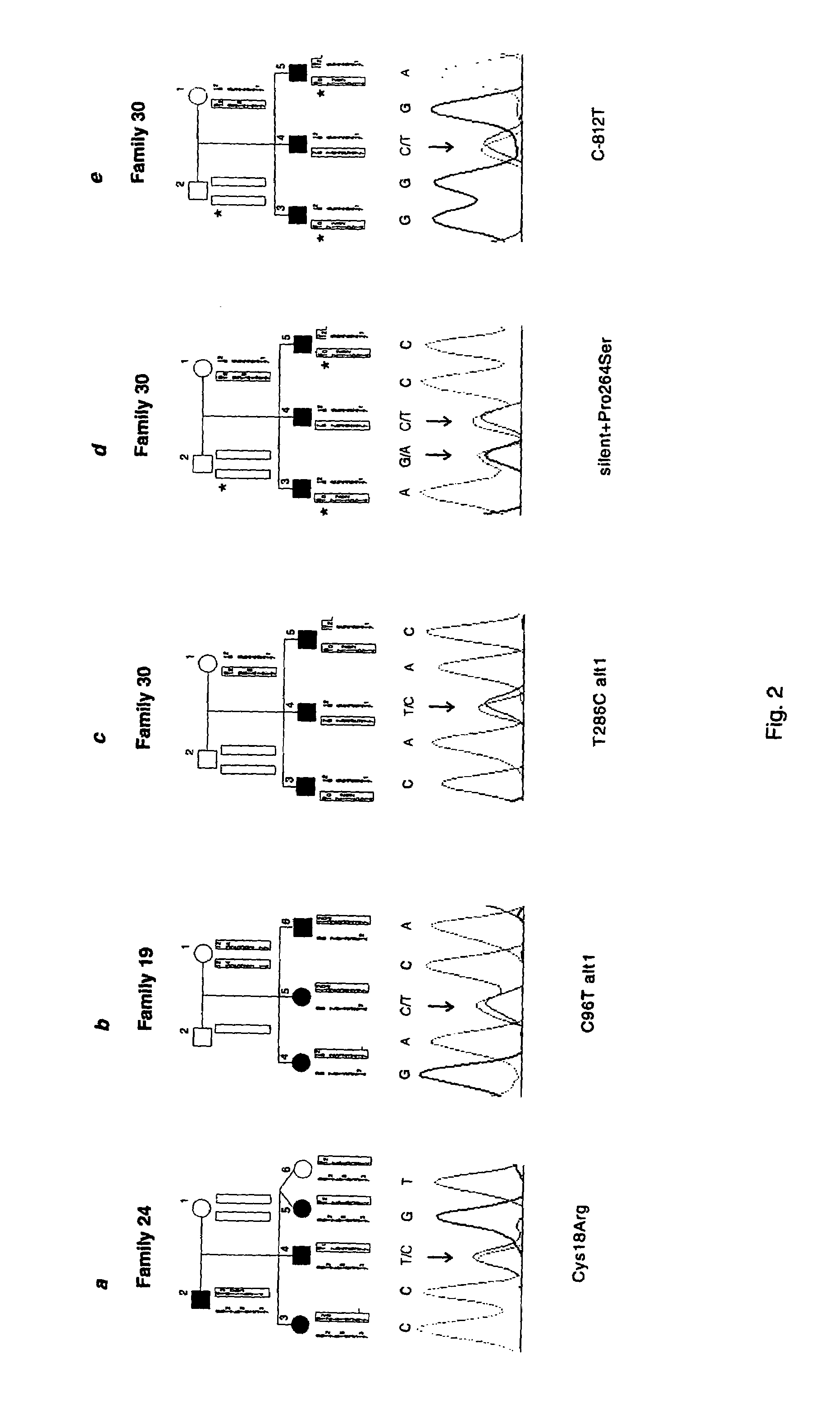
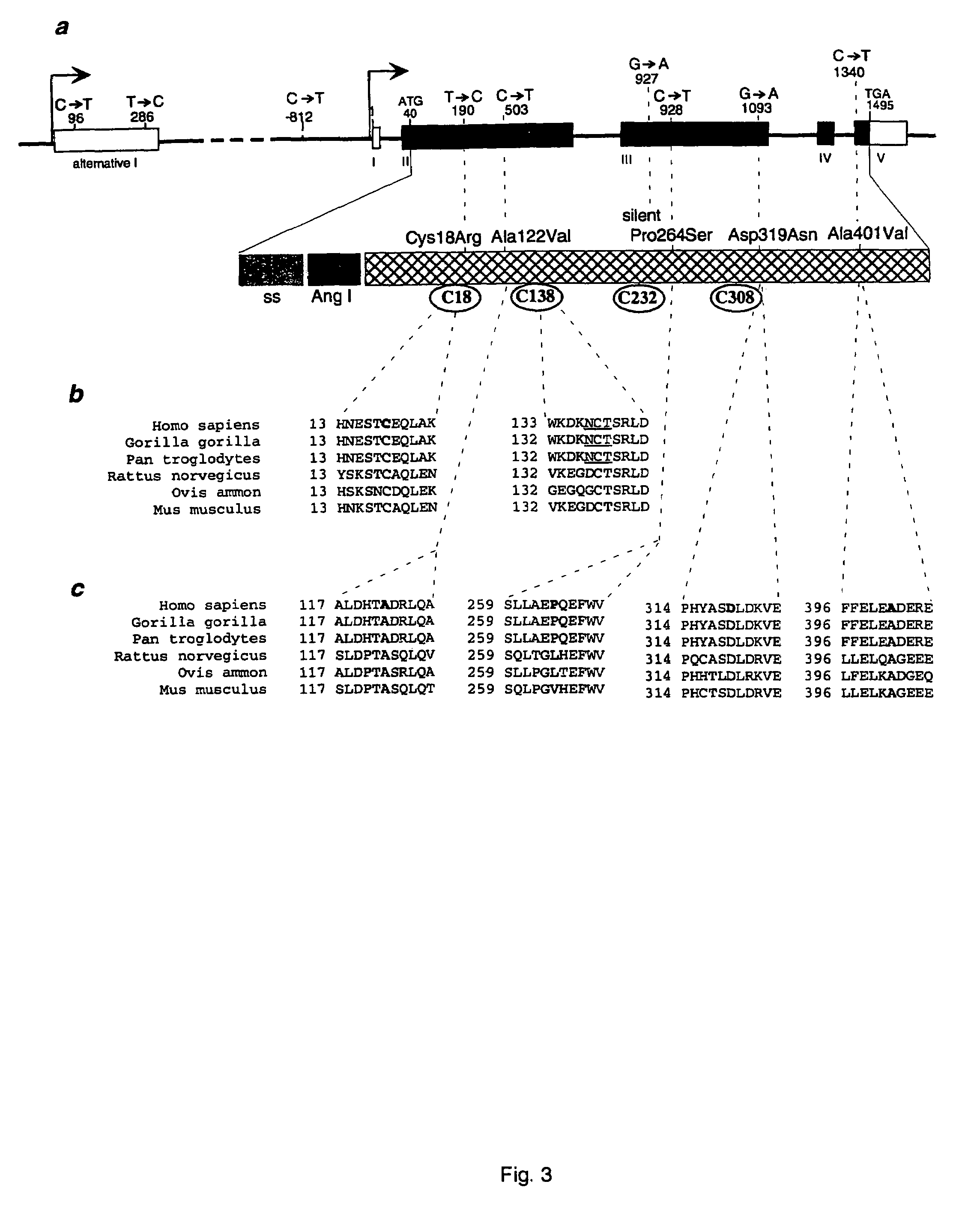
The present invention relates generally to the field of human genetics. Specifically, the present invention relates to methods and materials used to isolate and detect human diabetes mellitus predisposing gene, specifically the angiotensinogen (AGT) gene, some mutant alleles of which cause susceptibility to insulin-dependent diabetes mellitus (IDDM). More specifically, the invention relates to germline mutations in the AGT gene and their use in the diagnosis of predisposition to diabetes. The invention also relates to the prophylaxis and / or therapy of diabetes associated with a mutation in the AGT gene. The invention further relates to the screening of drugs for diabetes therapy. Finally, the invention relates to the screening of the AGT gene for mutations, which are useful for diagnosing the predisposition to diabetes.
Owner:MYRIAD GENETICS
Alterations in the dystrophin gene associated with sporadic dilated cardiomyopathy
ActiveUS7449561B1Preventing and treating sporadic DCMReduce and prevent gene expressionSugar derivativesMicrobiological testing/measurementGermline mutationMutant allele
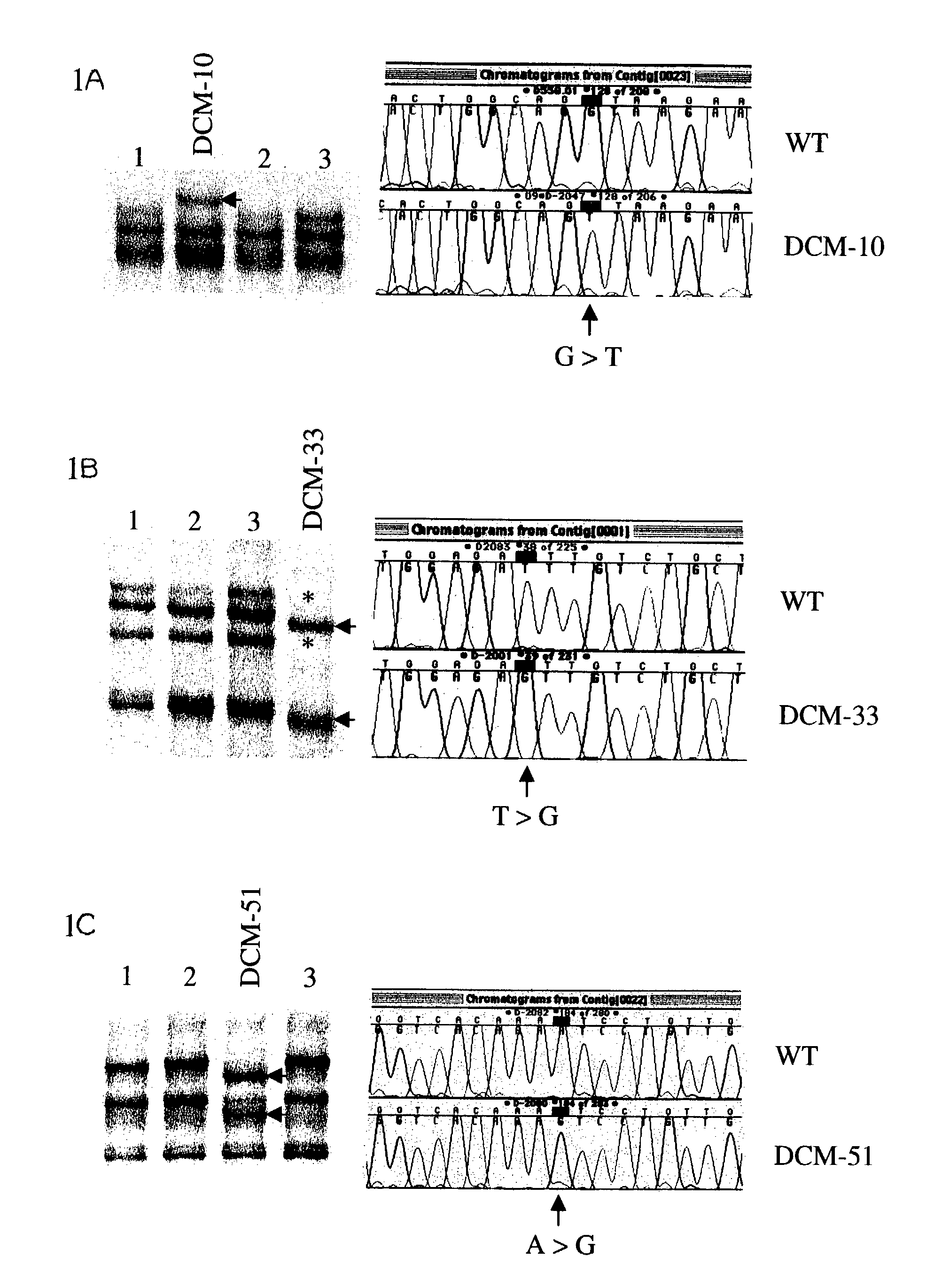
The present invention relates generally to the field of human genetics. Specifically, the present invention relates to methods and materials used to detect a human sporadic DCM predisposing gene, specifically the dystrophin gene, some mutant alleles of which cause susceptibility to sporadic DCM. More specifically, the invention relates to germline mutations in the dystrophin gene and their use in the diagnosis of predisposition to sporadic DCM. The invention also relates to the prophylaxis and / or therapy of sporadic DCM associated with a mutation in the dystrophin gene. The invention further relates to the screening of drugs for sporadic DCM therapy. Finally, the invention relates to the screening of the dystrophin gene for mutations / alterations, which are useful for diagnosing the predisposition to sporadic DCM.
Owner:CITY OF HOPE +2
Method for distinguishing somatic mutation and germline mutation
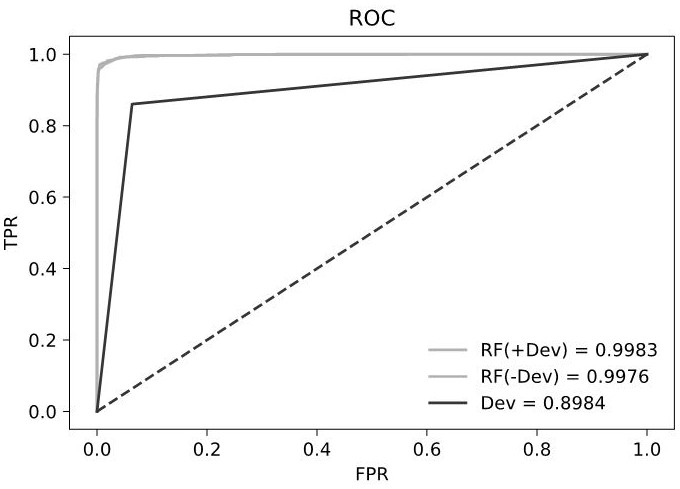
The invention relates to a method for distinguishing a somatic mutation and a germline mutation. The method comprises the following steps of obtaining at least one mutation site from a subject sample; obtaining a wild type support fragment and a mutant type support fragment, wherein the wild type support fragment is a cfDNA fragment containing a wild type base sequence, the mutant type support fragment is a cfDNA fragment containing a mutant type base sequence, the wild type base sequence is the same as a nucleotide sequence of a human reference genome at a corresponding position of a mutation site, and the mutant type base sequence is different; obtaining the number of the wild type support fragments with at least one length, obtaining the number of the corresponding mutant type support fragments with the same length, and calculating the difference value between the ratio of the wild type support fragments with the same length to the total number of the corresponding support fragments and the the ratio of the mutant type support fragments with the same length to the total number of the corresponding support fragments ; and taking the difference value as a distinguishing index. Method and device for identifying ctDNA from cfDNA are provided. The method is used for tumor family management and TMB detection.
Owner:GUANGZHOU BURNING ROCK DX CO LTD
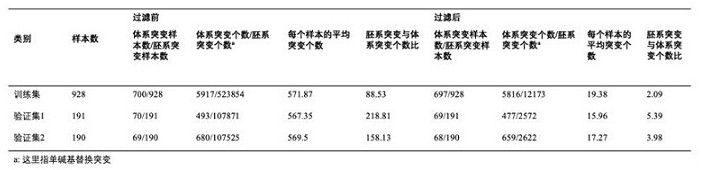
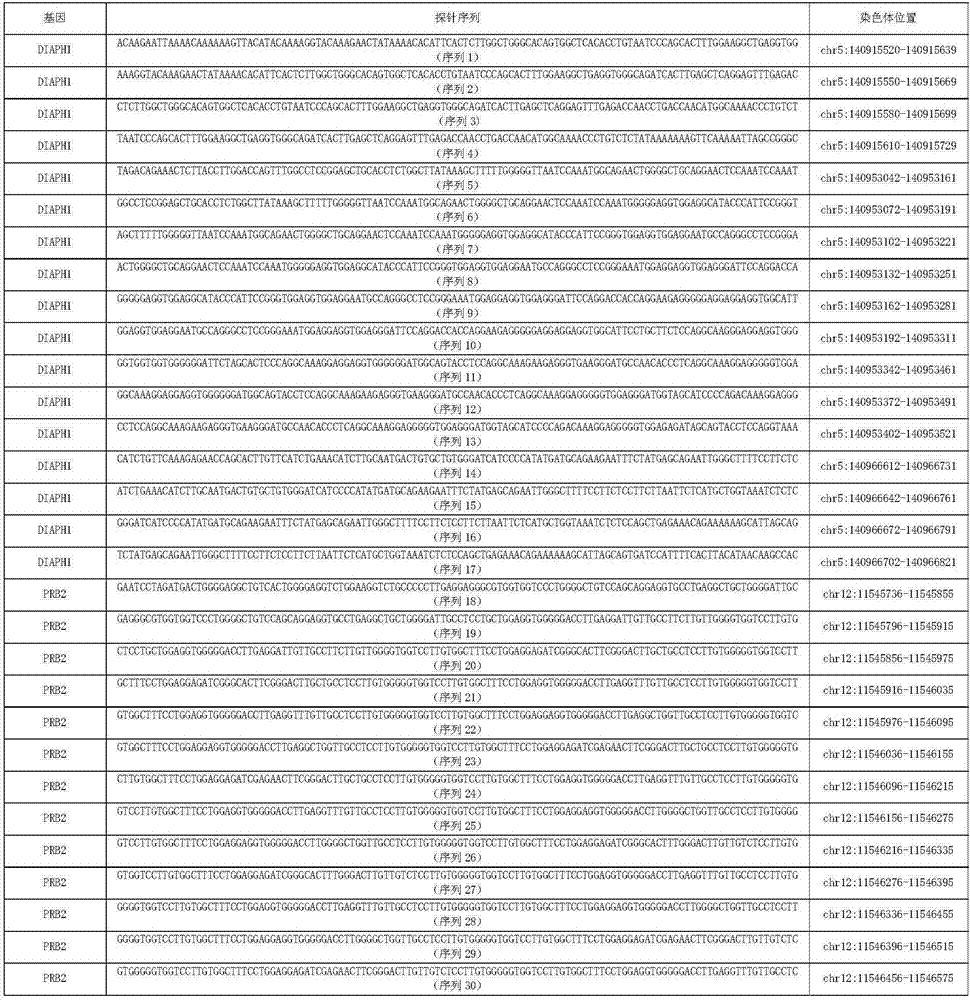
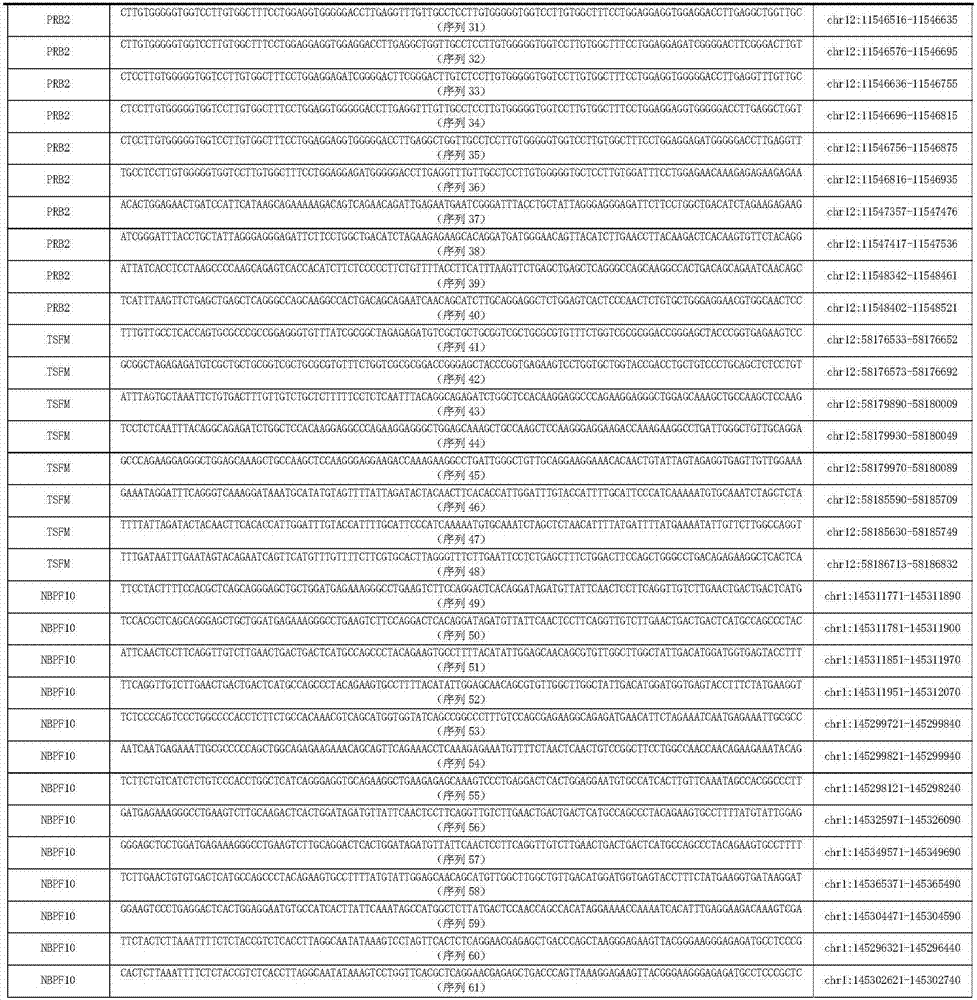
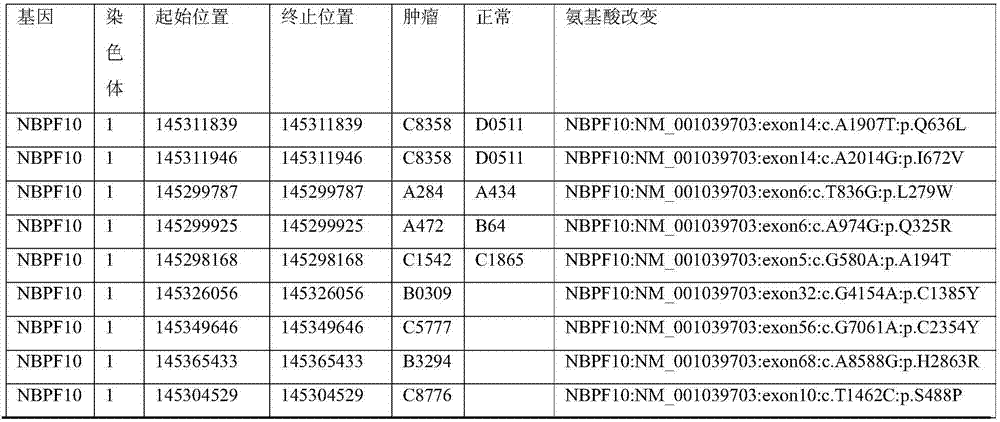
Kit for simultaneously detecting mutations of genes NBPF10, TSFM, PRB2 and DIAPH1
ActiveCN107974502AMicrobiological testing/measurementDNA/RNA fragmentationGermline mutationMRD Negative
The present invention discloses a kit for simultaneously detecting mutations of genes NBPF10, TSFM, PRB2 and DIAPH1, wherein the kit comprises a plurality of multi-gene capture probes, can be used fordetecting the somatic mutations and the germline mutations at the tumor genome level, can be used for the assisted diagnosis of patients with hematological tumors (especially acute lymphoblastic leukemia patients), the monitoring of minimal residual diseases, the evaluation of prognosis, and the like, and can provide important effect in the field of medical detection, wherein the detection methodis simple and sensitive.
Owner:PEOPLES HOSPITAL PEKING UNIV
Obesity gene and use thereof
InactiveUS7314713B1Susceptibility to obesityIncreased adipositySugar derivativesMicrobiological testing/measurementGermline mutationDiabetes mellitus
The present invention relates generally to the field of human genetics. Specifically, the present invention relates to methods and materials used to isolate and detect a human obesity and diabetes predisposing gene, specifically the TBC1D1 gene, some mutant alleles of which cause susceptibility to obesity and / or diabetes. More specifically, the invention relates to germline mutations in the TBC1D1 gene and their use in the diagnosis of predisposition to obesity and diabetes. Finally, the invention relates to the screening of the TBC1D1 gene for mutations / alterations, which are useful for diagnosing the predisposition to obesity.
Owner:UNIV OF UTAH RES FOUND
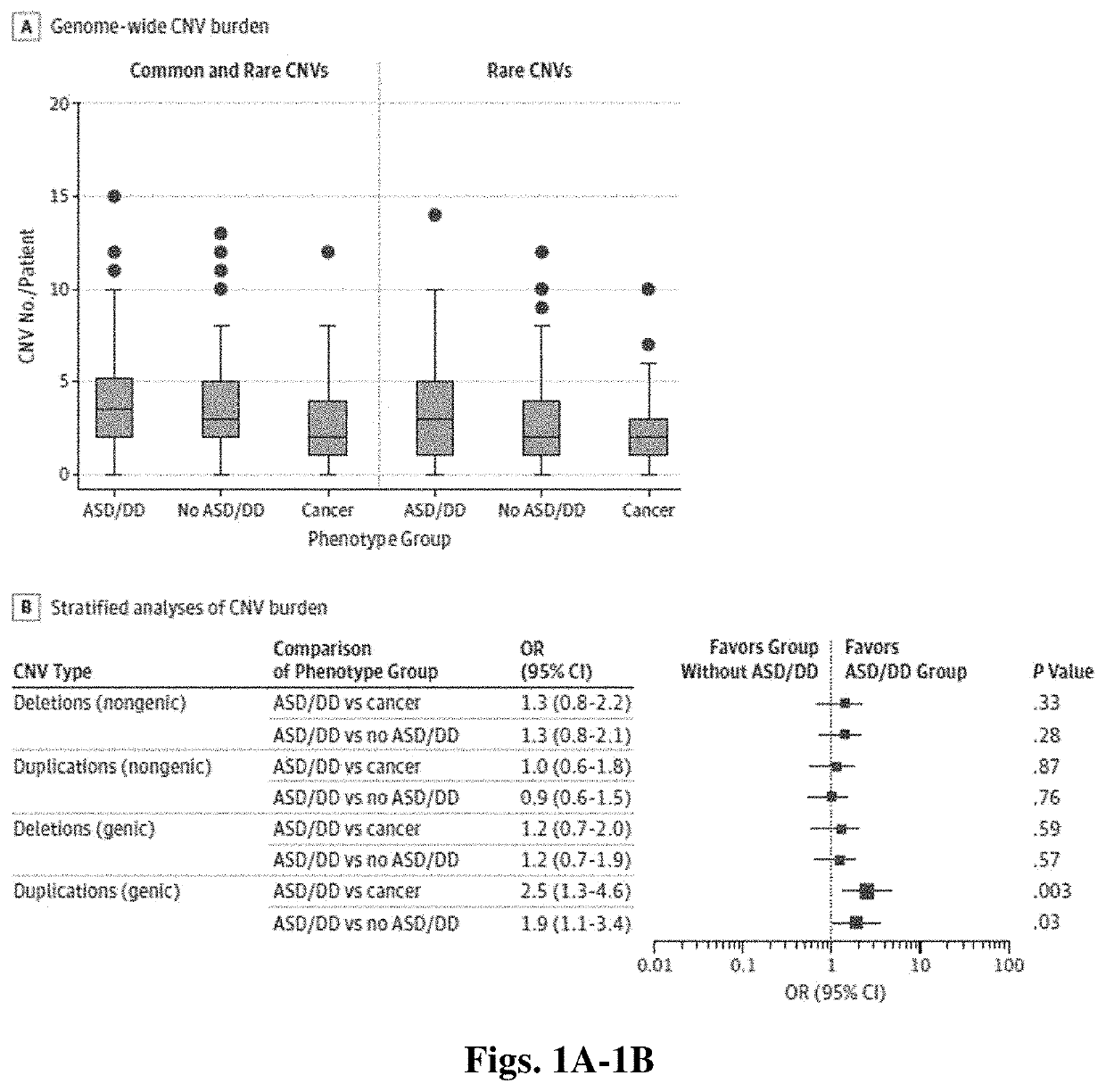
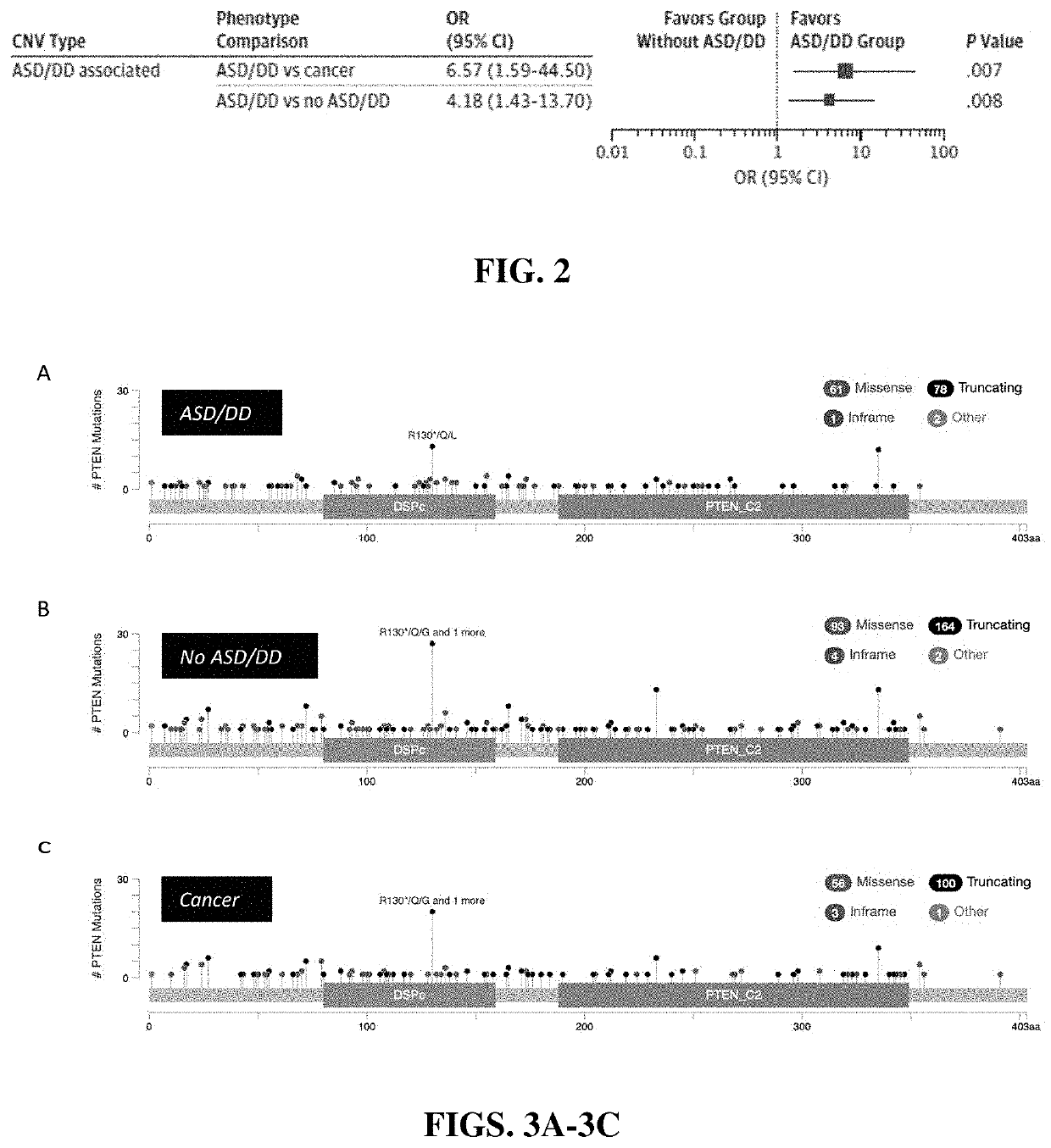
Distinguishing cancer from neurological risk using copy number variation
PendingUS20210190769A1Increased risk of developingHigh risk of developingMicrobiological testing/measurementDisease diagnosisGermline mutationNervous system
A method of providing a diagnosis for a subject having Cowden-like syndrome or PTEN germline mutations is described. The method includes the steps of: (a) obtaining a biological sample from a subject; (b) conducting a germline PTEN mutation and deletion analysis of genomic DNA from the biological sample; (c) determining the level of copy number variation in the genomic DNA; (d) comparing the level of copy number variation in the genomic DNA to a control value for copy number variation; and (e) diagnosing the subject as having an increased risk of developing a neurodevelopmental disorder if the copy number variation level is higher than the control value.
Owner:THE CLEVELAND CLINIC FOUND
Depression gene
InactiveUS20080160519A1Avoid depressionImprove disease symptomsSugar derivativesPeptide/protein ingredientsGermline mutationGenes mutation
The present invention relates generally to the field of human genetics. Specifically, the present invention relates to methods and materials used to isolate and detect a human depression predisposing gene, specifically the apoptotic protease activating factor 1 (APAF1) gene, some mutant alleles of which cause susceptibility to depression. More specifically, the invention relates to germline mutations in the APAF1 gene and their use in the diagnosis of predisposition to depression. The invention also relates to the prophylaxis and / or therapy of depression associated with a mutation in the APAF1 gene. The invention further relates to the screening of drugs for depression therapy. Finally, the invention relates to the screening of the APAF1 gene for mutations / alterations, which are useful for diagnosing the predisposition to depression.
Owner:IHC HEALTH SERVICES
Thalassemia mutant gene detection primer composition and reagent thereof
ActiveCN113981059ABetter ratioGood target rateMicrobiological testing/measurementDNA/RNA fragmentationForward primerGermline mutation
The invention discloses a thalassemia mutant gene detection primer composition. The thalassemia mutant gene detection primer composition comprises 131 pairs of first primers; each pair of first primers comprises a forward primer and a reverse primer, and each of the forward primer and the reverse primer comprises an amplification primer pair and other sequences connected with the 5' end of the amplification primer pair; the specific sequence of each pair of forward primer and reverse primer is selected from the sequences as shown in SEQ ID NO.1 to 262. According to the invention, three germline mutation related genes, namely HBA1 / 2 and HBB, which are important to thalassemia, are selected, not only can common SNV and short indel be detected, but also complex CNV mutation can be detected, and through peripheral blood gDNA detection, on one hand, genetic diagnosis of alpha and / or beta thalassemia can be carried out on suspected thalassemia patients, and on the other hand, genetic evaluation can be carried out on high-risk couples with thalassemia, and prenatal genetic diagnosis can be carried out on fetuses.
Owner:PILLAR BIOSCI INC
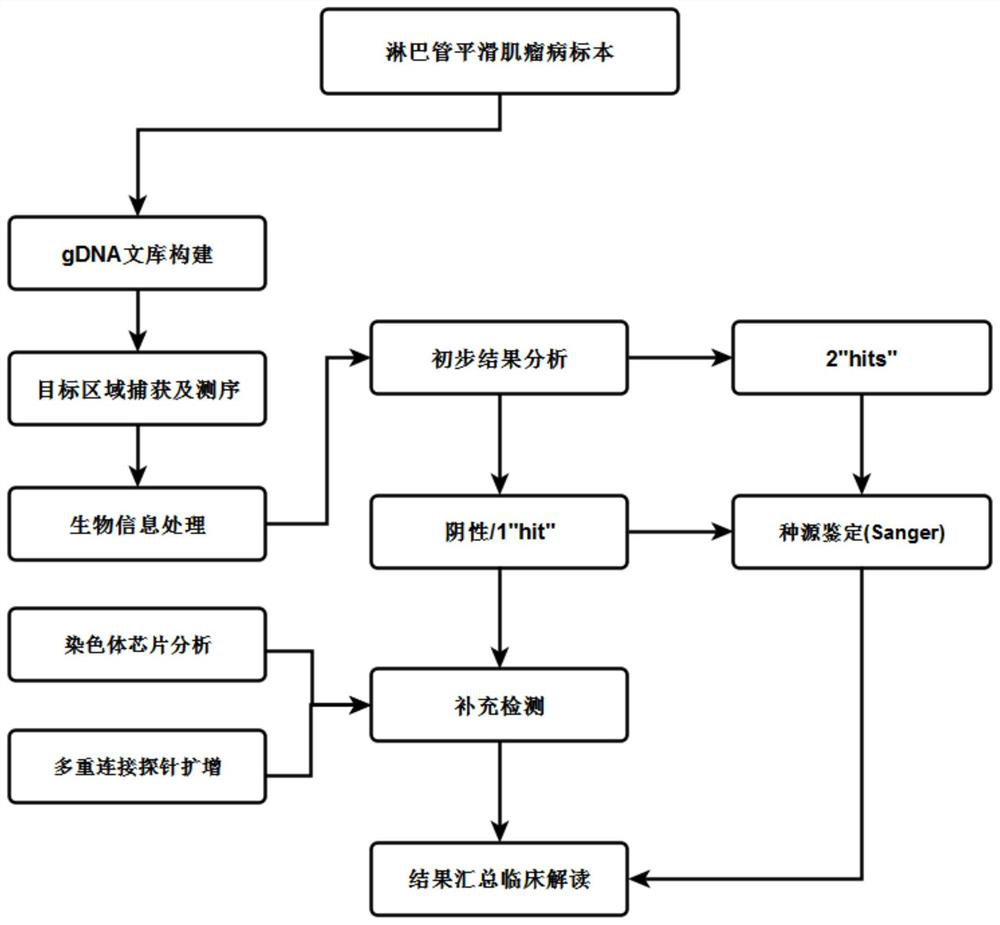
Combined detection method and application of lymphangioleiomyomatosis
ActiveCN110885879BImprove positive variant detection rateMicrobiological testing/measurementGermline mutationLymphatic vessel
The invention relates to a combined detection method for lymphangioleiomyomatosis and its application, and belongs to the technical field of gene detection. The method includes the following steps: liquid phase capture targeted sequencing: Panel design for the whole-exon coding regions of TSC1 and TSC2 genes highly related to LAM and mutant genes closely related to solid tumors, construction of gDNA library, and after hybridization capture, On-machine sequencing; sorting: analyze the above sequencing data by bioinformatics processing, if the TSC1 and TSC2 genes are detected negative, the chromosome chip analysis method is used for supplementary detection; if the single hit site is detected, the multiple ligation probe is used Amplification is used for supplementary testing; if loci with unclear somatic mutation and germline mutation origin are detected, Sanger test is used for supplementary testing. This method improves the detection rate of positive variants in LAM patients.
Owner:GUANGZHOU KINGMED DIAGNOSTICS GRP CO LTD
Use of tmem236 gene in the preparation of neoadjuvant radiotherapy resistance markers for diagnosing rectal cancer
ActiveCN114350813BImprove diagnostic accuracyStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationGermline mutationCancer research
The present invention provides the use of TMEM236 gene in preparing a marker for diagnosing locally advanced rectal cancer neoadjuvant radiotherapy primary resistance. The TMEM236 gene mutation site is chr10:17796115. If it is detected that the TMEM236 gene is at the site The point mutation of chr10:17796115, TMEM236:NM_001098844:exon4:c.G667A, shows that the patient is resistant to radiotherapy. The present invention also provides the use of the kit for detecting TMEM236 gene mutation in the preparation of a neoadjuvant radiotherapy resistance diagnostic composition for diagnosing locally advanced rectal cancer. The germline mutation mutant gene TMEM236 screened by the invention can be used for predicting the primary resistance of patients with locally advanced rectal cancer to neoadjuvant radiotherapy.
Owner:广州达安临床检验中心有限公司 +2
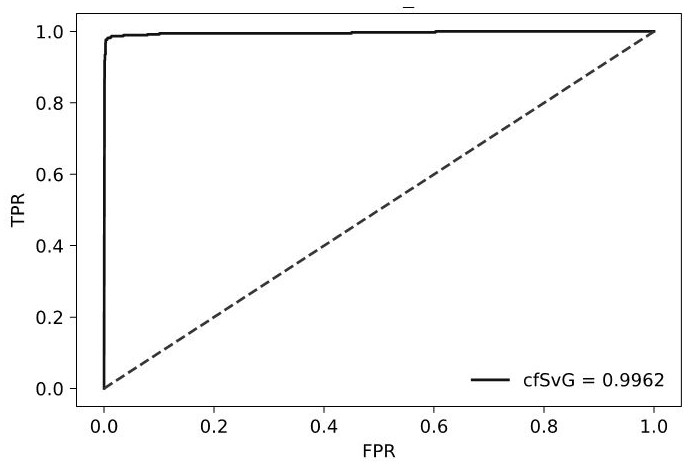
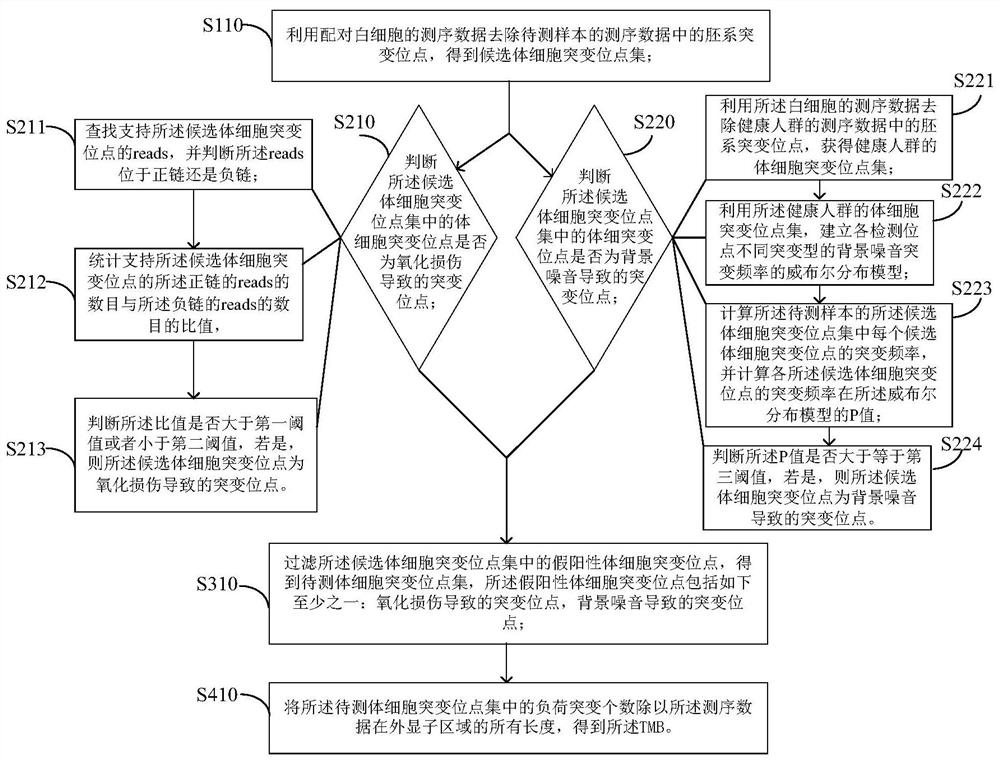
Method and device for detecting tmb
ActiveCN110689930BImprove accuracyImprove stabilityBiostatisticsProteomicsGermline mutationWhite blood cell
The invention provides a method and device for detecting TMB. The method comprises: using the sequencing data of paired white blood cells to remove the germline mutation sites in the sequencing data of the sample to be tested to obtain a set of candidate somatic mutation sites; filtering false positive somatic mutation sites in the set of candidate somatic mutation sites to obtain The set of somatic mutation sites to be detected, the false positive somatic mutation sites include at least one of the following: mutation sites caused by oxidative damage, mutation sites caused by background noise; load mutations that concentrate the somatic mutation sites to be detected The number is divided by all the lengths of the sequencing data in the exon region to obtain the TMB. The accuracy and stability of TMB are improved by making full use of paired leukocytes, background noise mutation frequency distribution database, and oxidative damage to remove false positive somatic mutations.
Owner:北京橡鑫生物科技有限公司 +2
med12 gene mutation detection kit and its application
ActiveCN110229897BThe detection method is simpleSensitive detection methodMicrobiological testing/measurementDNA/RNA fragmentationGermline mutationMyeloid leukemia
The invention discloses a MED12 gene mutation detection kit and an application thereof. The kit of the present invention includes a probe set for detecting MED12 gene mutation, and the probe set is composed of probe 1-probe 48. The kit of the present invention can not only be used to detect MED12 somatic mutation and germline mutation at the level of tumor genome, but also can be used for auxiliary diagnosis, monitoring of minimal residual disease and evaluation of prognosis in patients with hematological tumors, especially acute myeloid leukemia patients. , and the detection method of the kit of the present invention is simple, sensitive and accurate, and will play an important role in the field of medical detection.
Owner:PEOPLES HOSPITAL PEKING UNIV
A method for differentiating gene mutation types from individual tumor samples based on next-generation sequencing
ActiveCN110846411BLow detection costImprove detection efficiencyMicrobiological testing/measurementProteomicsGermline mutationSomatic cell
The invention relates to a method for distinguishing gene mutation types an individual tumor sample based on second-generation sequencing. A tumor tissue sample and a normal tissue sample are used forlibrary construction and NGS sequencing respectively, strand bias and different types of base frequencies of mutation sites stored in an intermediate file BAM for biological information analysis of the tumor tissue sample are analyzed, the quality of base comparison and the frequency of noise are used as the training characteristics of machine learning, meanwhile, type information of corresponding mutation sites of the normal tissue sample is paired to serve as a prediction mutation type, a classification prediction model is constructed to distinguish somatic mutation from germline mutation,the model is used to distinguish somatic mutation from germline mutation, the detection efficiency is high, the specificity is high, and after the model is established, the individual tumor sample canbe used for NGS sequencing and mutation detection, the detection cost of a normal or cancer sample can be well saved, and meanwhile, the problem that normal tissues of tumor patients with specific types are difficult to obtain can be solved.
Owner:上海仁东医学检验所有限公司 +1
Sequencing and analysis of exosome associated nucleic acids
ActiveUS11396676B2Microbiological testing/measurementPreparing sample for investigationGermline mutationNucleotide
The invention provides a series of steps that prepare nucleic acids (RNA and / or DNA) isolated from extracellular vesicles for sequencing. This enables a wide diversity of RNAs and / or DNAs, to be efficiently detected. These can then be used to identify various attributes such as gene expression, alternative splicing, and the detection of both somatic and germline mutations including single nucleotide variants (SNV) and structural variations (insertions / deletions, fusions, inversions).
Owner:EXOSOME DIAGNOSTICS
Obesity gene and use thereof
InactiveUS8034564B2Prevent obesityImprove disease symptomsSugar derivativesMicrobiological testing/measurementGermline mutationDiabetes mellitus
The present invention relates generally to the field of human genetics. Specifically, the present invention relates to methods and materials used to isolate and detect a human obesity and diabetes predisposing gene, specifically the TBC1D1 gene, some mutant alleles of which cause susceptibility to obesity and / or diabetes. More specifically, the invention relates to germline mutations in the TBC1D1 gene and their use in the diagnosis of predisposition to obesity and diabetes. Finally, the invention relates to the screening of the TBC1D1 gene for mutations / alterations, which are useful for diagnosing the predisposition to obesity.
Owner:UNIV OF UTAH RES FOUND
Determining a predisposition to cancer
The present invention relates to methods and kits for determining a predisposition for developing cancer, e.g., prostate and / or breast cancer, due to a germline mutation of a NBS1 gene. The present invention also relates to surveillance protocols for developing cancer, e.g., prostate and / or breast cancer, due to germline mutation of a NBS1 gene.
Owner:POMERANIAN ACAD OF MEDICINE
Mono-allelic mutation analysis for identifying germline mutations
A diagnostic strategy for detection of inherited diseases caused by germline mutations is based on somatic cell hybridization. Each allele of a human gene involved in the inherited disease is isolated in a somatic cell hybrid. The products of the isolated human allele are then observed in the absence of the other allele of the human.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com