Method for distinguishing gene mutation type from individual tumor sample based on second-generation sequencing
A next-generation sequencing and sample technology, applied in the fields of genomics, sequence analysis, biochemical equipment and methods, etc., can solve the problem of not taking into account the positive and negative strand-biased germline mutation frequency characteristics of sequenced DNA fragments, and determine as somatic cells. Mutation, low specificity of differentiation and judgment, etc., to achieve the effect of saving detection costs, high specificity, and high detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0023] The present invention will be described in detail below in combination with specific embodiments and accompanying drawings.
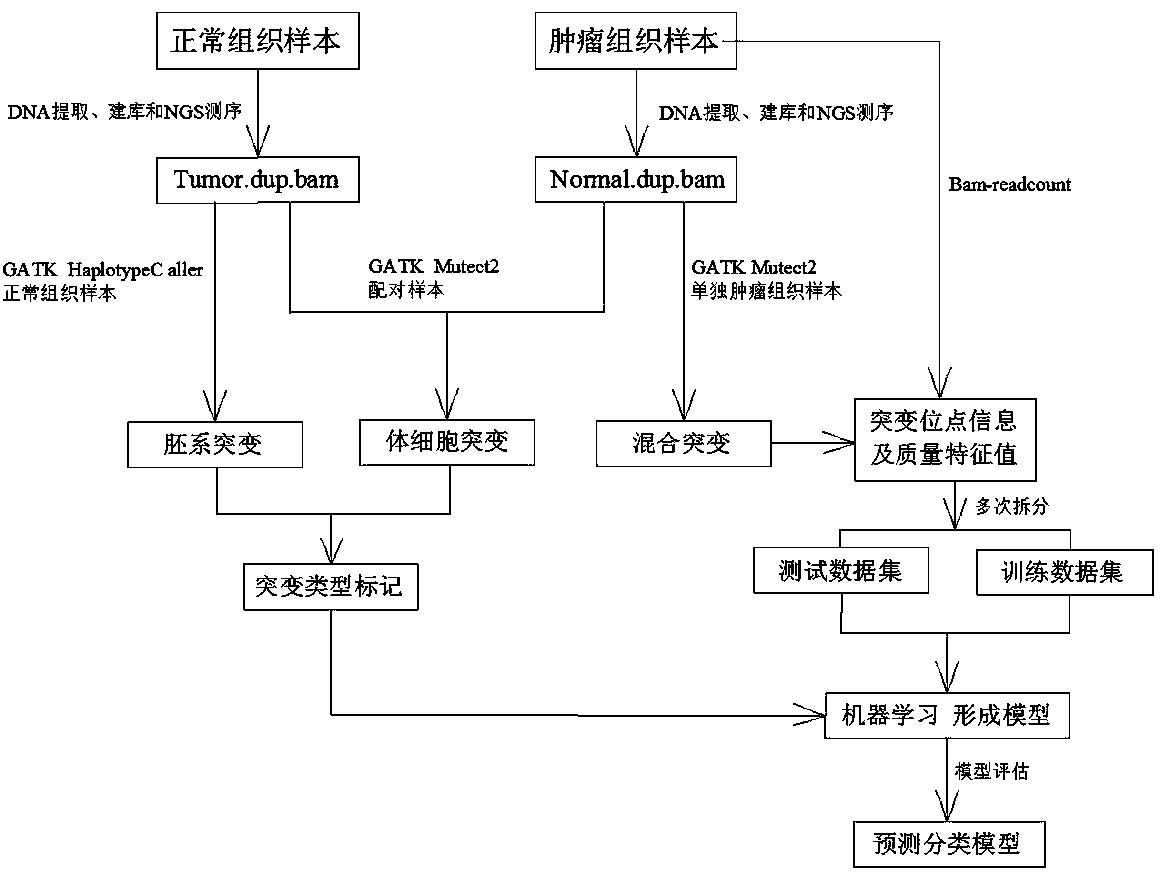
[0024] See attached figure 1 , a method for distinguishing gene mutation types based on next-generation sequencing of individual tumor samples, specifically comprising the following steps:
[0025] S1. Select 189 target regions, extract DNA from 189 target regions in tumor tissue samples and normal tissue samples, and use the method of probe capture to build and sequence the DNA extracted from tumor tissue samples and normal tissue samples , to obtain the raw data of 189 target region sequencing;
[0026] S2. Use the BWA MEM algorithm to perform sequence comparison on the DNA sequencing data from tumor tissue samples and normal tissue samples respectively obtained through step S1, and simultaneously generate a comparison file Tumor.dup.bam belonging to tumor tissue samples and a comparison file belonging to normal tissues Sample comparison file...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com