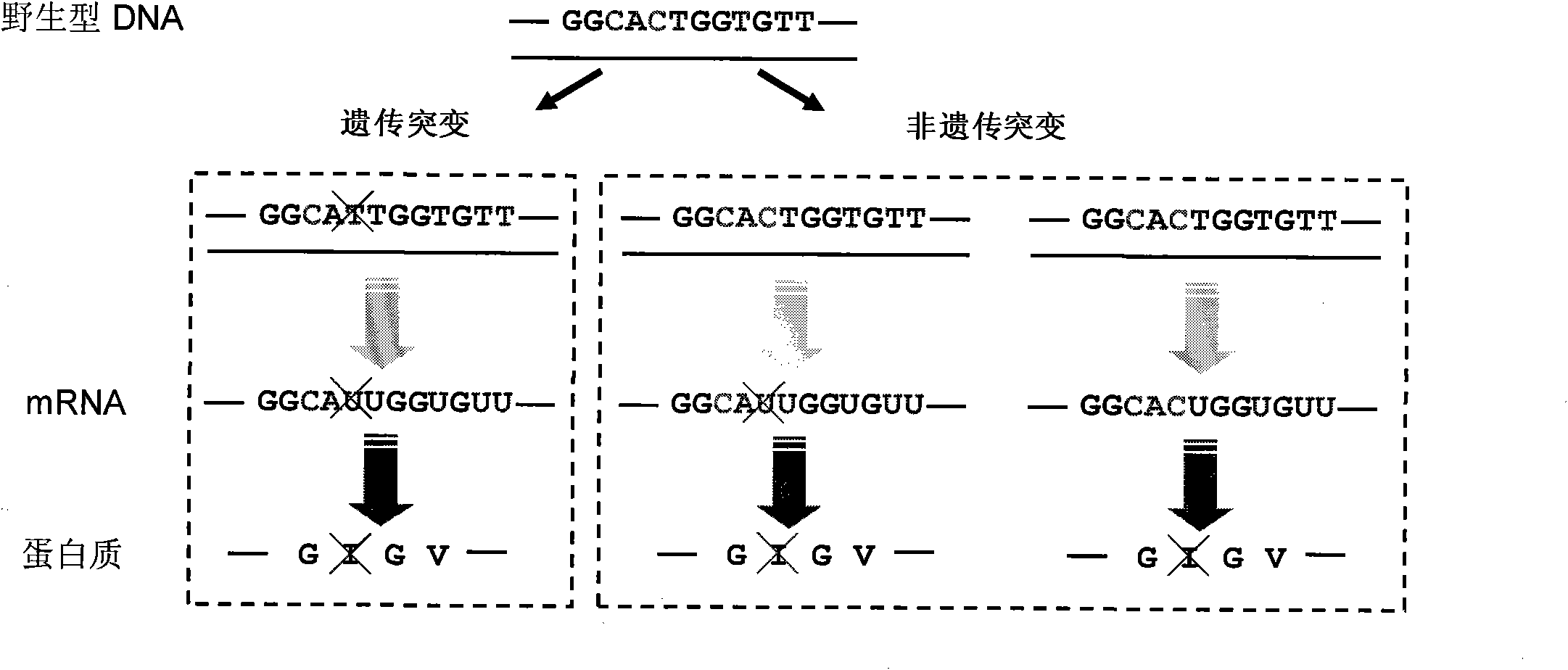
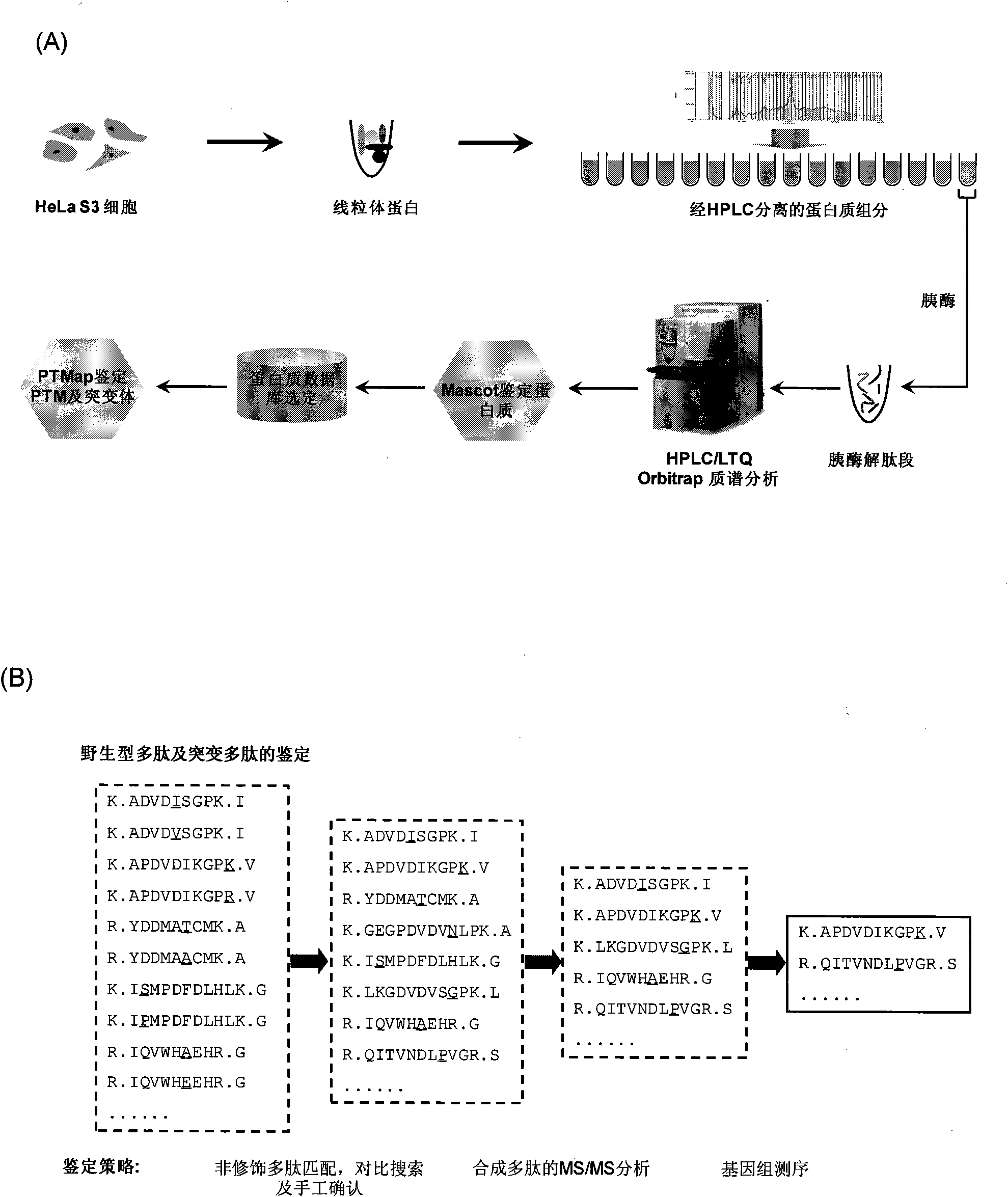
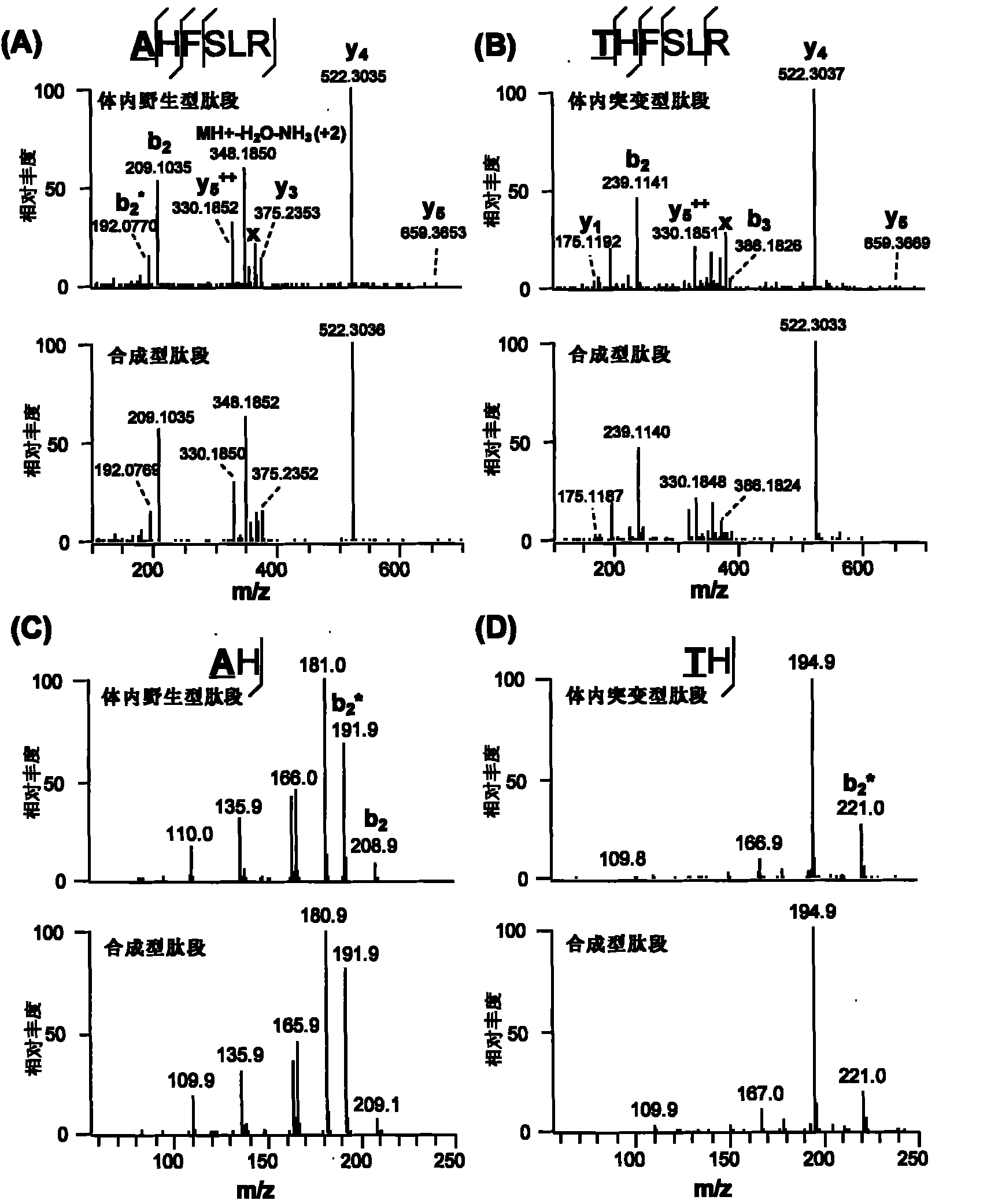
Identification method of highly stoichiometric non-germline mutation in eucaryotic cell
An identification method and eukaryotic cell technology, applied in the field of identification of non-hereditary mutations with high stoichiometry in eukaryotic cells, can solve the problems of typical high error rate and insufficient reliability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0014] Materials and Methods
[0015] Preparation of Mitochondria from Eukaryotic Cells Using Discontinuous Gradient Separation Method (22,23) Mitochondria were isolated from HeLa S3 cells (Biovest International Inc. Minneapolis, MN). All equipment and solutions were cooled on ice during preparation. The preparation process is briefly described as follows: resuspend 50 ml of cell pellet in 500 ml of hypotonic buffer supplemented with protease inhibitors (Sigma, St.Louis, MO) for 20 minutes (10 mM Tris pH 7.3, 1.5 mM MgCl 2 , 10mM KCl, 10mM 2-Mercaptoethanol); after homogenization, centrifuge at 1000g for 10 minutes; collect the supernatant and centrifuge at 20,000g for 30 minutes; resuspend the centrifuged pellet in 6ml of HIM buffer (200mM manna Alcohol mannitol, 70mM sucrose sucrose, 1mM EGTA, 10mM HEPES, pH 7.5, containing protease inhibitors), then transferred to ultracentrifuge tubes containing 70% / 20% (v / v) Percoll solution (GE Healthcare, Piscataway, NJ) Centrifuge a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com