Method and device for detecting tmb
A technology of sequencing data and mutation sites, applied in genomics, instrumentation, sequence analysis, etc., can solve problems such as inaccurate TMB detection, and achieve the effect of improving accuracy and stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
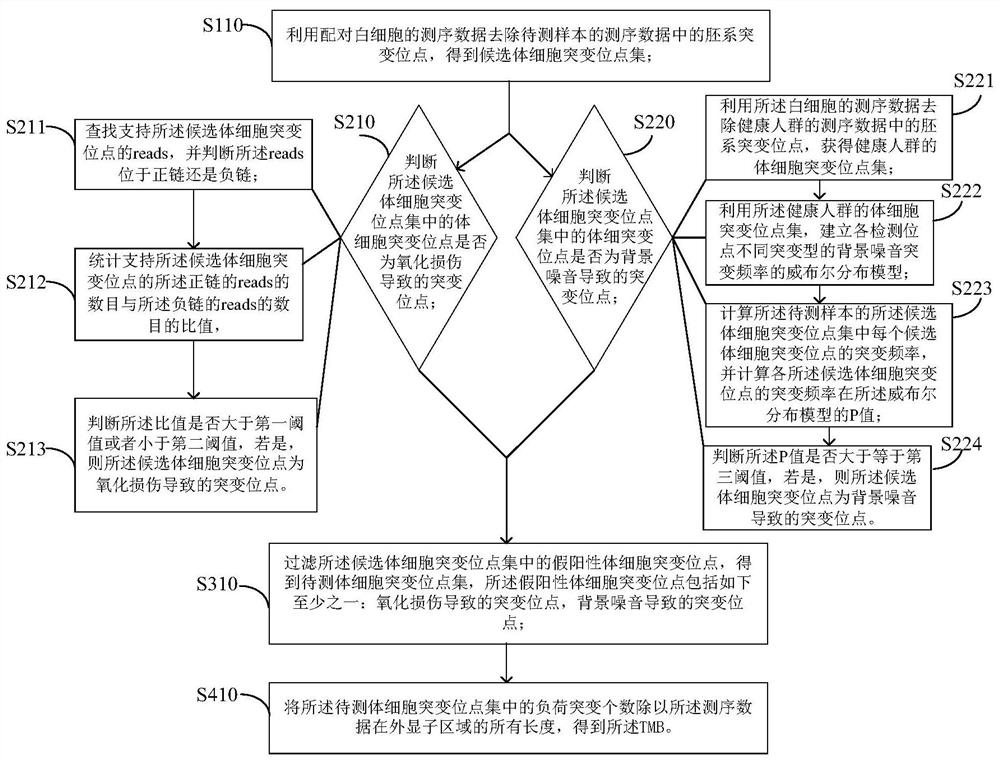
[0052] The present application provides an embodiment of a method for detecting TMB.
[0053] figure 1 is a flow chart of an optional method for detecting TMB according to an embodiment of the present invention, such as figure 1 As shown, the method includes:
[0054] Step S101, using the sequencing data of paired white blood cells to remove the germline mutation sites in the sequencing data of the sample to be tested to obtain a set of candidate somatic mutation sites;
[0055] Step S102, filtering the false positive somatic mutation sites in the set of candidate somatic mutation sites to obtain the set of somatic mutation sites to be tested, the false positive somatic mutation sites include at least one of the following: mutation sites caused by oxidative damage , the mutation site caused by background noise;
[0056] Step S103, dividing the number of mutation loads in the concentration of somatic mutation sites to be detected by all the lengths of the sequencing data in ...
Embodiment 2
[0084] Taking a clinical sample as an example, obtain the patient's tissue sample and its corresponding plasma sample, extract the DNA and build the database, and use the illumina sequencing platform to obtain the off-machine data sampA.R1.fastq.gz, sampA.R2 of the tissue respectively. fastq.gz; and its white blood cell data are sampB.R1.fastq.gz, sampB.R2.fastq.gz; after bwa comparison, marker duplication and base quality correction, sampA.final.bam and sampB are obtained. final. bam. Use sampA.final.bam and sampB.final.bam to detect samples according to the pairing mode of mutect2, and obtain sampA.result.vcf. Use sampA.final.bam to detect according to the non-pairing mode of mutect2, and get sampA.resultWithoutSampB.vcf. According to the filter module and calculation module described in the patent, the subsequent filtering and calculation of sampA.resultvcf is performed, and the final TMB value is 14.7; if the existing method is used to detect the module, the pairing mode ...
Embodiment 3
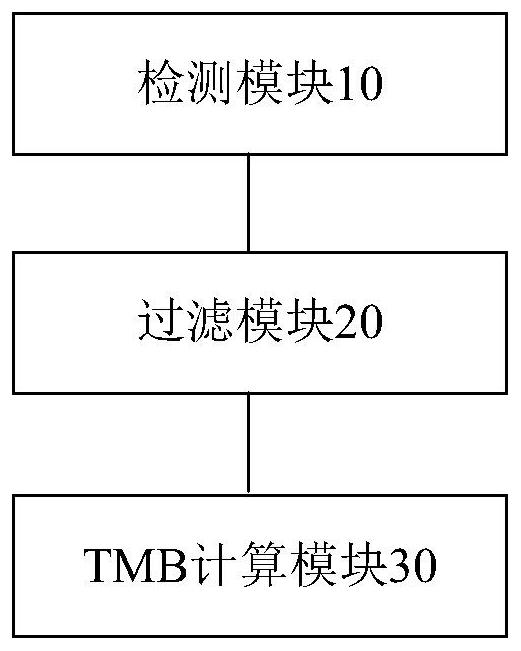
[0090] The present application also provides an embodiment of a device for detecting TMB.
[0091] image 3 is a schematic diagram of an optional device for detecting TMB according to an embodiment of the present invention, such as image 3 As shown, the device includes a detection module 10, a filtering module 20, and a TMB calculation module 30. The detection module 10 is used to remove germline mutation sites in the sequencing data of the sample to be tested by using the sequencing data of paired white blood cells to obtain candidate somatic cells. Mutation site set; filtering module 20, used to filter false positive somatic mutation sites in the set of candidate somatic mutation sites to obtain a set of somatic mutation sites to be tested. False positive somatic mutation sites include at least one of the following : mutation sites caused by oxidative damage, mutation sites caused by background noise; TMB calculation module 30, used to divide the number of load mutations i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com