Method for obtaining stable DNA tetrahedron synthesis parameters based on modeling
An acquisition method and tetrahedron technology are applied in the field of acquisition of stable DNA tetrahedron synthesis parameters, which can solve the problems such as the inability to improve the space capacity of DNA tetrahedrons, and achieve the effects of not cumbersome design, wide application and low cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
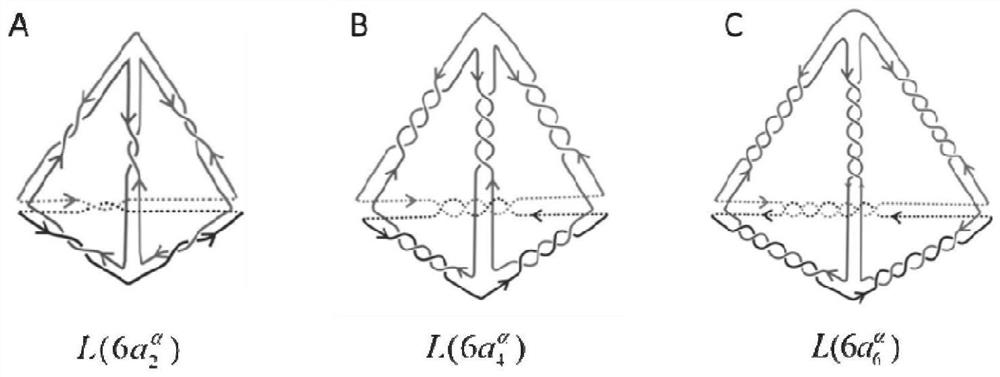
[0080] (1) Using CorelDRAW 12 software to draw a mathematical chain link with specific topological properties figure 1 TD, such as figure 1 As shown, its link graph contains 4 branches, and the number of intersections on each edge is 2.
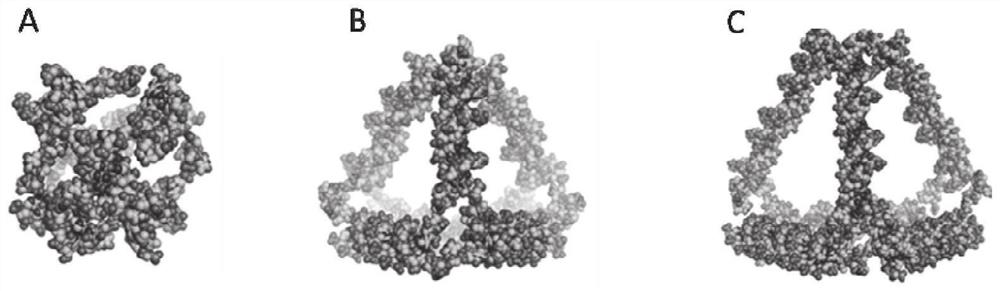
[0081](2) According to the chain link diagram, use the PolygenDNA model software to set the apex connector to be composed of 5 thymidines. By adjusting the module parameters in the program, construct a DNA tetrahedron nanocage all-atom model, such as figure 2 As shown, name it 1TD.pdb.
[0082] Wherein: the double helix edge torsion number of the DNA tetrahedral cage model obtained by constructing is respectively 1, corresponds to the crossing number of the mathematical link diagram in the step (1), and its all-atom model structure atom number is respectively 4586, constitutes a tetrahedron The number of nucleotides in the structure is 144, which is a standard DNA tetrahedron all-atom structure.
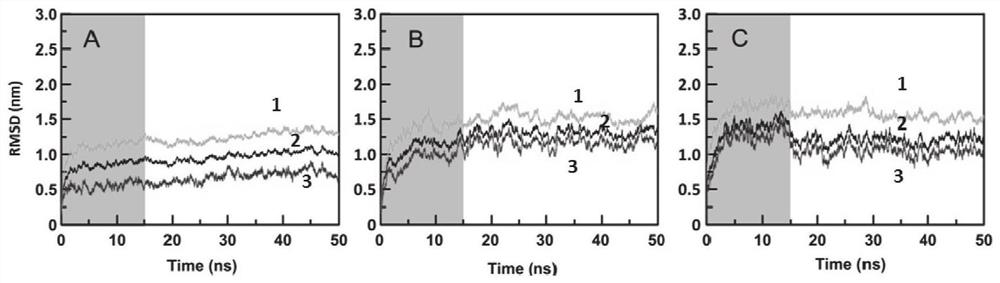
[0083] (3) Using Gromacs 5.1.2 software, ...
Embodiment 2
[0096] (1) Using CorelDRAW 12 software to draw a mathematical chain link with specific topological properties figure 1 TD, such as figure 1 As shown, its link graph contains 4 branches, and the number of crossings on each edge is 4.
[0097] (2) According to the chain link diagram, use the PolygenDNA model software to set the apex connector to be composed of 5 thymidines. By adjusting the module parameters in the program, construct a DNA tetrahedron nanocage all-atom model, such as figure 2 As shown, name it 2TD.pdb.
[0098] Wherein: the double helix edge torsion number of the DNA tetrahedral cage model obtained by constructing is respectively 2, corresponds to the intersection number of the mathematical link diagram in the step (1), and its all-atom model structure atom number is respectively 8393, constitutes a tetrahedron The number of nucleotides in the structure is 264, which is a standard DNA tetrahedron all-atom structure.
[0099] (3) Using Gromacs 5.1.2 software,...
Embodiment 3
[0112] (1) Using CorelDRAW 12 software to draw a mathematical chain link with specific topological propertiesfigure 1 TD, such as figure 1 As shown, its chain-ring graph contains 4 branches, and the number of crossings on each edge is 6.
[0113] (2) According to the chain link diagram, use the PolygenDNA model software to set the apex connector to be composed of 5 thymidines. By adjusting the module parameters in the program, construct a DNA tetrahedron nanocage all-atom model, such as figure 2 As shown, name it 3TD.pdb.
[0114] Wherein: the double helix edge torsion number of the DNA tetrahedral cage model obtained by constructing is respectively 3, and corresponding to the intersection number of the mathematical link diagram in the step (1), its all-atom model structure atom number is respectively 11825, constitutes a tetrahedron The number of nucleotides in the structure is 372, which is a standard DNA tetrahedron all-atom structure.
[0115] (3) Using Gromacs 5.1.2 so...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com